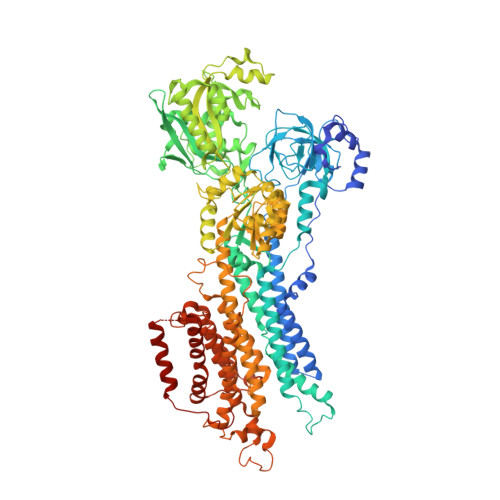
Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Zhang, Y., Inoue, M., Tsutsumi, A., Watanabe, S., Nishizawa, T., Nagata, K., Kikkawa, M., Inaba, K.(2020) Sci Adv 6: eabb0147-eabb0147
- PubMed: 32851169
- DOI: https://doi.org/10.1126/sciadv.abb0147
- Primary Citation of Related Structures:
6LLE, 6LLY, 6LN5, 6LN6, 6LN7, 6LN8, 6LN9 - PubMed Abstract:
Sarco/endoplasmic reticulum Ca 2+ ATPase (SERCA) pumps Ca 2+ from the cytosol into the ER and maintains the cellular calcium homeostasis. Herein, we present cryo-electron microscopy (cryo-EM) structures of human SERCA2b in E1∙2Ca 2+ -adenylyl methylenediphosphonate (AMPPCP) and E2-BeF 3 - states at 2.9- and 2.8-Å resolutions, respectively. The structures revealed that the luminal extension tail (LE) characteristic of SERCA2b runs parallel to the lipid-water boundary near the luminal ends of transmembrane (TM) helices TM10 and TM7 and approaches the luminal loop flanked by TM7 and TM8. While the LE served to stabilize the cytosolic and TM domain arrangement of SERCA2b, deletion of the LE rendered the overall conformation resemble that of SERCA1a and SERCA2a and allowed multiple conformations. Thus, the LE appears to play a critical role in conformational regulation in SERCA2b, which likely explains the different kinetic properties of SERCA2b from those of other isoforms lacking the LE.
- Institute of Multidisciplinary Research for Advanced Materials, Tohoku University, Sendai 980-8577, Japan.
Organizational Affiliation: