Structural Insights into the Trans -Acting Enoyl Reductase in the Biosynthesis of Long-Chain Polyunsaturated Fatty Acids in Shewanella piezotolerans .
Zhang, M., Zhang, H., Li, Q., Gao, Y., Guo, L., He, L., Zang, S., Guo, X., Huang, J., Li, L.(2021) J Agric Food Chem 69: 2316-2324
- PubMed: 33587627
- DOI: https://doi.org/10.1021/acs.jafc.0c07386
- Primary Citation of Related Structures:
6LKC - PubMed Abstract:
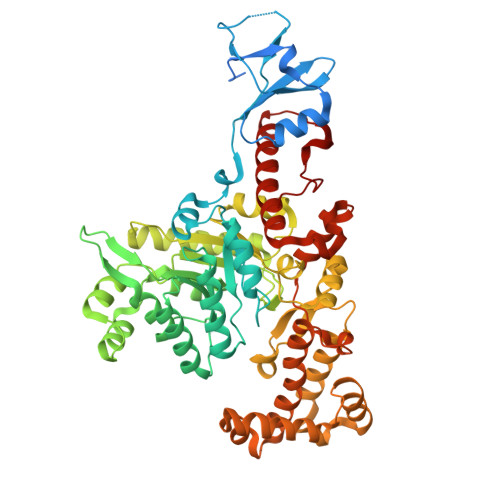
Two long-chain polyunsaturated fatty acids (LC-PUFAs), eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA), play vital roles in human health. Similarly, two biosynthetic pathways, based on desaturase/elongase and polyketide synthase, have been implicated in the synthesis of microbial LC-PUFA. Up to now, only several microalgae, no bacteria, have been used in the commercial production of oils rich in DHA and/or EPA. Fully understanding the enzymatic mechanism in the biosynthesis of LC-PUFA would contribute significantly to produce EPA and/or DHA by the bacteria. In this study, we report a 1.998 Å-resolution crystal structure of trans -acting enoyl reductase (ER), SpPfaD, from Shewanella piezotolerans . The SpPfaD model consists of one homodimer in the asymmetric unit, and each subunit contains three domains. These include an N-terminal, a central domain forming a classic TIM barrel with a single FMN cofactor molecule bound atop the barrel, and a C-terminal domain with a lid above the TIM barrel. Furthermore, we docked oxidized nicotinamide adenine dinucleotide phosphate (NADP) and an inhibitor 2-(4-(2-((3-(5-(pyridin-2-ylthio)thiazol-2-yl)ureido)methyl)-1 H -imidazole-4-yl)phenoxy)acetic acid (TUI) molecule into the active site and analyzed the inhibition and catalytic mechanisms of the enoyl reductase SpPfaD. To the best of our knowledge, this is the first crystal structure of trans -ER in the biosynthesis of bacterial polyketides.
- Engineering Research Center of Industrial Microbiology, Ministry of Education; Collaborative Innovation Center of Haixi Green Bio-Manufacturing Technology, Ministry of Education; National & Local Joint Engineering Research Center of Industrial Microbiology and Fermentation Technology, National Development and Reform Commission; College of Life Sciences, Fujian Normal University, Fuzhou, 350117, P. R. China.
Organizational Affiliation: