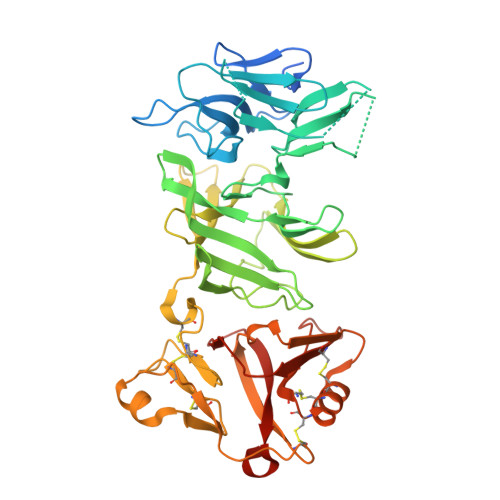
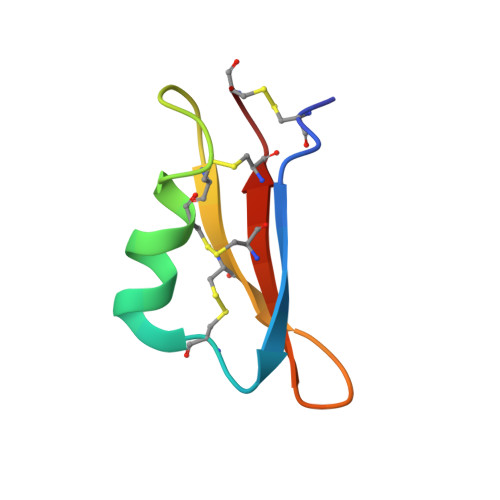
Mechanism of self/nonself-discrimination in Brassica self-incompatibility.
Murase, K., Moriwaki, Y., Mori, T., Liu, X., Masaka, C., Takada, Y., Maesaki, R., Mishima, M., Fujii, S., Hirano, Y., Kawabe, Z., Nagata, K., Terada, T., Suzuki, G., Watanabe, M., Shimizu, K., Hakoshima, T., Takayama, S.(2020) Nat Commun 11: 4916-4916
- PubMed: 33004803
- DOI: https://doi.org/10.1038/s41467-020-18698-w
- Primary Citation of Related Structures:
6KYW - PubMed Abstract:
Self-incompatibility (SI) is a breeding system that promotes cross-fertilization. In Brassica, pollen rejection is induced by a haplotype-specific interaction between pistil determinant SRK (S receptor kinase) and pollen determinant SP11 (S-locus Protein 11, also named SCR) from the S-locus. Although the structure of the B. rapa S 9 -SRK ectodomain (eSRK) and S 9 -SP11 complex has been determined, it remains unclear how SRK discriminates self- and nonself-SP11. Here, we uncover the detailed mechanism of self/nonself-discrimination in Brassica SI by determining the S 8 -eSRK-S 8 -SP11 crystal structure and performing molecular dynamics (MD) simulations. Comprehensive binding analysis of eSRK and SP11 structures reveals that the binding free energies are most stable for cognate eSRK-SP11 combinations. Residue-based contribution analysis suggests that the modes of eSRK-SP11 interactions differ between intra- and inter-subgroup (a group of phylogenetically neighboring haplotypes) combinations. Our data establish a model of self/nonself-discrimination in Brassica SI.
- Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo, 113-8657, Japan.
Organizational Affiliation: