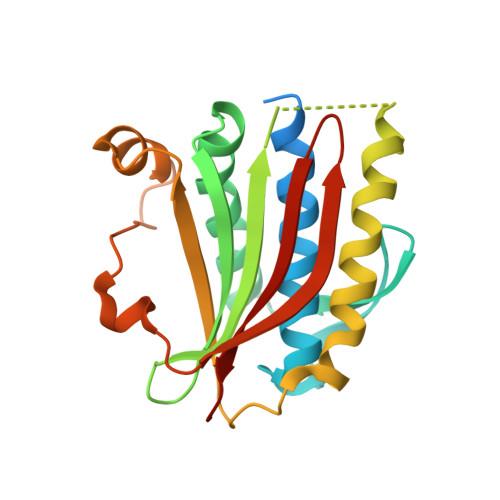
Molecular basis for assembly of the shieldin complex and its implications for NHEJ.
Liang, L., Feng, J., Zuo, P., Yang, J., Lu, Y., Yin, Y.(2020) Nat Commun 11: 1972-1972
- PubMed: 32332881
- DOI: https://doi.org/10.1038/s41467-020-15879-5
- Primary Citation of Related Structures:
6KTO - PubMed Abstract:
Shieldin, including SHLD1, SHLD2, SHLD3 and REV7, functions as a bridge linking 53BP1-RIF1 and single-strand DNA to suppress the DNA termini nucleolytic resection during non-homologous end joining (NHEJ). However, the mechanism of shieldin assembly remains unclear. Here we present the crystal structure of the SHLD3-REV7-SHLD2 ternary complex and reveal an unexpected C (closed)-REV7-O (open)-REV7 conformational dimer mediated by SHLD3. We show that SHLD2 interacts with O-REV7 and the N-terminus of SHLD3 by forming β sheet sandwich. Disruption of the REV7 conformational dimer abolishes the assembly of shieldin and impairs NHEJ efficiency. The conserved FXPWFP motif of SHLD3 binds to C-REV7 and blocks its binding to REV1, which excludes shieldin from the REV1/Pol ζ translesion synthesis (TLS) complex. Our study reveals the molecular architecture of shieldin assembly, elucidates the structural basis of the REV7 conformational dimer, and provides mechanistic insight into orchestration between TLS and NHEJ.
- Institute of Systems Biomedicine, Department of Pathology, Beijing Key Laboratory of Tumor Systems Biology, School of Basic Medical Sciences, Peking University Health Science Center, Beijing, 100191, China. liangling@hsc.pku.edu.cn.
Organizational Affiliation: