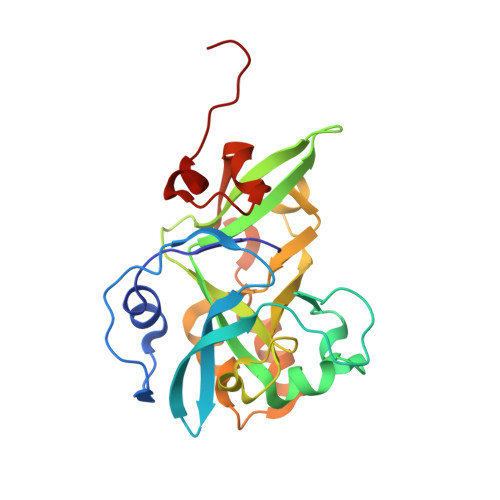
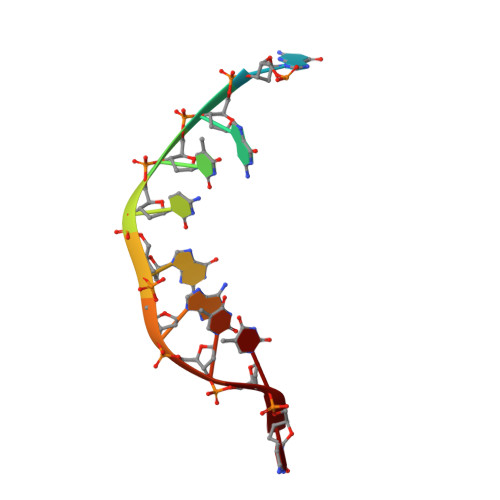
Molecular basis of abasic site sensing in single-stranded DNA by the SRAP domain of E. coli yedK.
Wang, N., Bao, H., Chen, L., Liu, Y., Li, Y., Wu, B., Huang, H.(2019) Nucleic Acids Res 47: 10388-10399
- PubMed: 31504793
- DOI: https://doi.org/10.1093/nar/gkz744
- Primary Citation of Related Structures:
6KBS, 6KBU, 6KBX, 6KBZ, 6KCQ, 6KIJ - PubMed Abstract:
HMCES and yedK were recently identified as sensors of abasic sites in ssDNA. In this study, we present multiple crystal structures captured in the apo-, nonspecific-substrate-binding, specific-substrate-binding, and product-binding states of yedK. In combination with biochemical data, we unveil the molecular basis of AP site sensing in ssDNA by yedK. Our results indicate that yedK has a strong preference for AP site-containing ssDNA over native ssDNA and that the conserved Glu105 residue is important for identifying AP sites in ssDNA. Moreover, our results reveal that a thiazolidine linkage is formed between yedK and AP sites in ssDNA, with the residues that stabilize the thiazolidine linkage important for the formation of DNA-protein crosslinks between yedK and the AP sites. We propose that our findings offer a unique platform to develop yedK and other SRAP domain-containing proteins as tools for detecting abasic sites in vitro and in vivo.
- Department of Biology, Southern University of Science and Technology, Shenzhen, Guangdong 518055, China.
Organizational Affiliation: