Inactivity of YGL082W in vitro due to impairment of conformational change in the catalytic center loop
Lu, L., Guo, Y., Wang, T., Liang, L., Zhao, S., Wang, F., Liu, L.(2019) Sci China B Chem
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| pseudo deubiquitinase | 283 | Saccharomyces cerevisiae S288C | Mutation(s): 0 Gene Names: YGL082W | 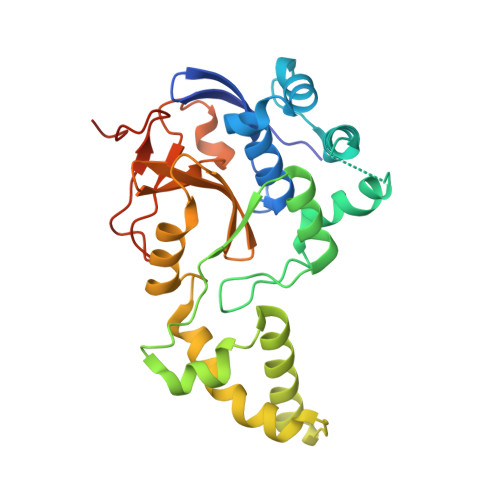 | |
UniProt | |||||
Find proteins for P53155 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P53155 Go to UniProtKB: P53155 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P53155 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 60.94 | α = 90 |
| b = 70.64 | β = 102.54 |
| c = 68.38 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| XDS | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Science Foundation (China) | China | -- |