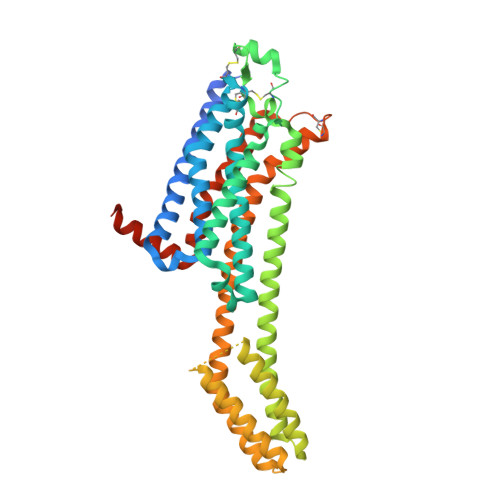
High-viscosity sample-injection device for serial femtosecond crystallography at atmospheric pressure.
Shimazu, Y., Tono, K., Tanaka, T., Yamanaka, Y., Nakane, T., Mori, C., Terakado Kimura, K., Fujiwara, T., Sugahara, M., Tanaka, R., Doak, R.B., Shimamura, T., Iwata, S., Nango, E., Yabashi, M.(2019) J Appl Crystallogr 52: 1280-1288
- PubMed: 31798359
- DOI: https://doi.org/10.1107/S1600576719012846
- Primary Citation of Related Structures:
6JZH, 6JZI - PubMed Abstract:
A sample-injection device has been developed at SPring-8 Angstrom Compact Free-Electron Laser (SACLA) for serial femtosecond crystallography (SFX) at atmospheric pressure. Microcrystals embedded in a highly viscous carrier are stably delivered from a capillary nozzle with the aid of a coaxial gas flow and a suction device. The cartridge-type sample reservoir is easily replaceable and facilitates sample reloading or exchange. The reservoir is positioned in a cooling jacket with a temperature-regulated water flow, which is useful to prevent drastic changes in the sample temperature during data collection. This work demonstrates that the injector successfully worked in SFX of the human A 2A adenosine receptor complexed with an antagonist, ZM241385, in lipidic cubic phase and for hen egg-white lysozyme microcrystals in a grease carrier. The injection device has also been applied to many kinds of proteins, not only for static structural analyses but also for dynamics studies using pump-probe techniques.
- RIKEN SPring-8 Center, 1-1-1 Kouto, Sayo-cho, Sayo-gun, Hyogo 679-5148, Japan.
Organizational Affiliation: