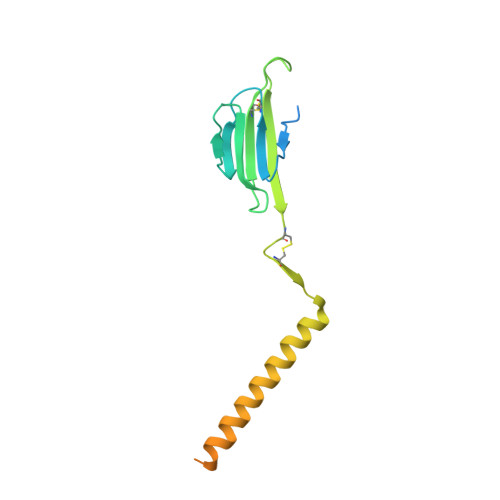
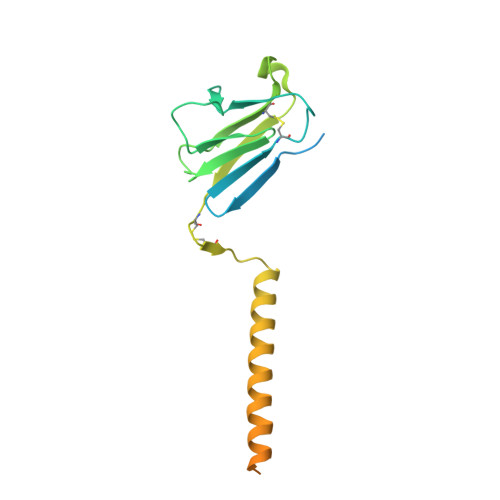
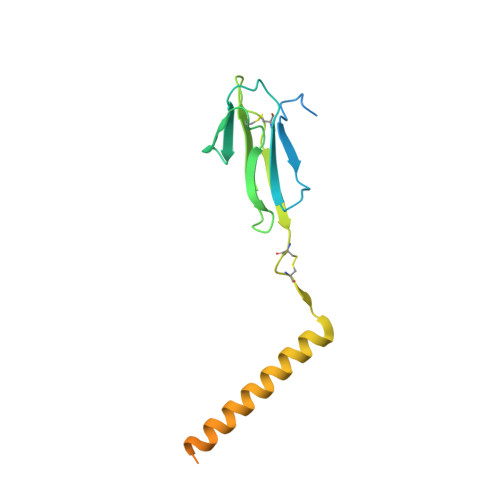
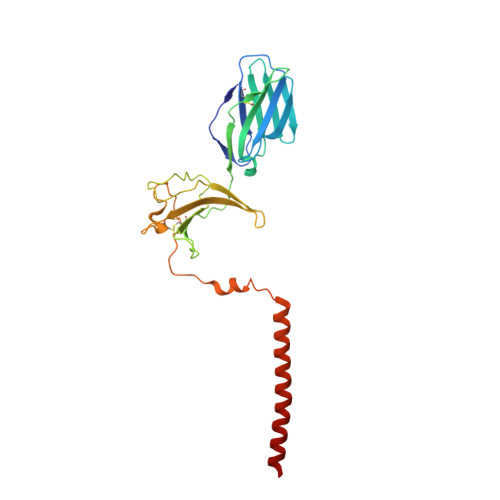
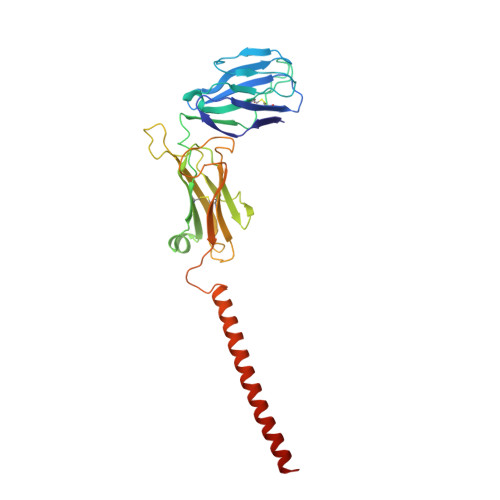
Structural basis of assembly of the human T cell receptor-CD3 complex.
Zheng, L., Lin, J., Zhang, B., Zhu, Y., Li, N., Xie, S., Wang, Y., Gao, N., Huang, Z.(2019) Nature 573: 546-552
- PubMed: 31461748
- DOI: https://doi.org/10.1038/s41586-019-1537-0
- Primary Citation of Related Structures:
6JXR - PubMed Abstract:
The αβ T cell receptor (TCR), in association with the CD3γε-CD3δε-CD3ζζ signalling hexamer, is the primary determinant of T cell development and activation, and of immune responses to foreign antigens. The mechanism of assembly of the TCR-CD3 complex remains unknown. Here we report a cryo-electron microscopy structure of human TCRαβ in complex with the CD3 hexamer at 3.7 Å resolution. The structure contains the complete extracellular domains and all the transmembrane helices of TCR-CD3. The octameric TCR-CD3 complex is assembled with 1:1:1:1 stoichiometry of TCRαβ:CD3γε:CD3δε:CD3ζζ. Assembly of the extracellular domains of TCR-CD3 is mediated by the constant domains and connecting peptides of TCRαβ that pack against CD3γε-CD3δε, forming a trimer-like structure proximal to the plasma membrane. The transmembrane segment of the CD3 complex adopts a barrel-like structure formed by interaction of the two transmembrane helices of CD3ζζ with those of CD3γε and CD3δε. Insertion of the transmembrane helices of TCRαβ into the barrel-like structure via both hydrophobic and ionic interactions results in transmembrane assembly of the TCR-CD3 complex. Together, our data reveal the structural basis for TCR-CD3 complex assembly, providing clues to TCR triggering and a foundation for rational design of immunotherapies that target the complex.
- HIT Center for Life Sciences, School of Life Science and Technology, Harbin Institute of Technology, Harbin, China.
Organizational Affiliation: