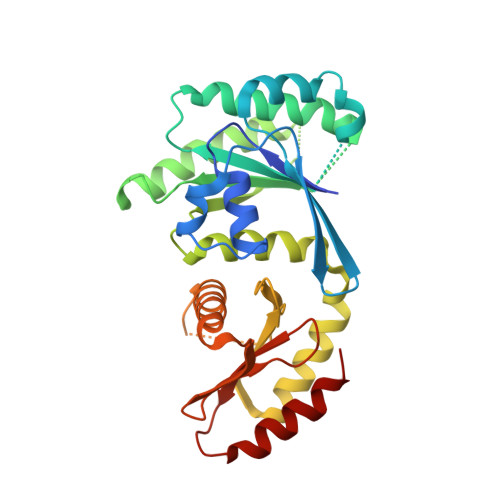
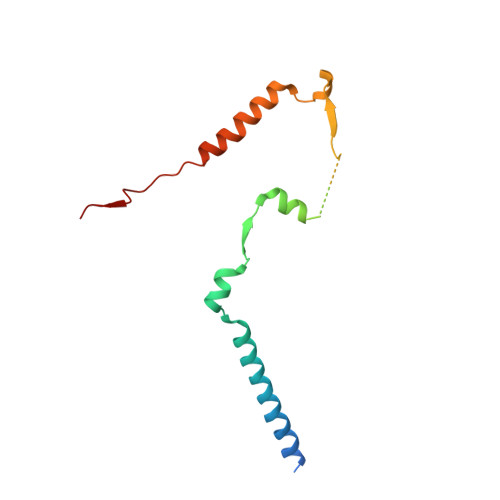
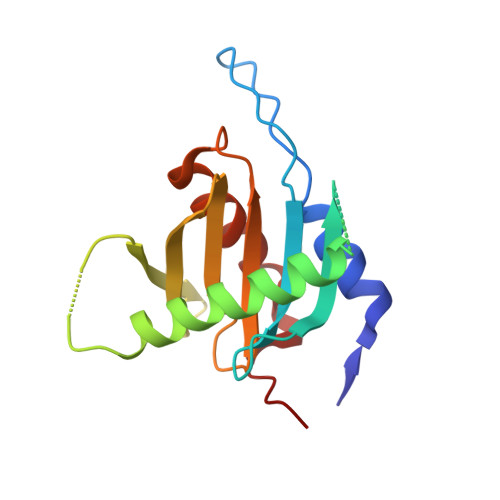
Structural insights into the EGO-TC-mediated membrane tethering of the TORC1-regulatory Rag GTPases.
Zhang, T., Peli-Gulli, M.P., Zhang, Z., Tang, X., Ye, J., De Virgilio, C., Ding, J.(2019) Sci Adv 5: eaax8164-eaax8164
- PubMed: 31579828
- DOI: https://doi.org/10.1126/sciadv.aax8164
- Primary Citation of Related Structures:
6JWP - PubMed Abstract:
The Rag/Gtr GTPases serve as a central module in the nutrient-sensing signaling network upstream of TORC1. In yeast, the anchoring of Gtr1-Gtr2 to membranes depends on the Ego1-Ego2-Ego3 ternary complex (EGO-TC), resulting in an EGO-TC-Gtr1-Gtr2 complex (EGOC). EGO-TC and human Ragulator share no obvious sequence similarities and also differ in their composition with respect to the number of known subunits, which raises the question of how the EGO-TC fulfills its function in recruiting Gtr1-Gtr2. Here, we report the structure of EGOC, in which Ego1 wraps around Ego2, Ego3, and Gtr1-Gtr2. In addition, Ego3 interacts with Gtr1-Gtr2 to stabilize the complex. The functional roles of key residues involved in the assembly are validated by in vivo assays. Our structural and functional data combined demonstrate that EGOC and Ragulator-Rag complex are structurally conserved and that EGO-TC is essential and sufficient to recruit Gtr1-Gtr2 to membranes to ensure appropriate TORC1 signaling.
- State Key Laboratory of Molecular Biology, CAS Center for Excellence in Molecular Cell Science, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, University of Chinese Academy of Sciences, Chinese Academy of Sciences, 320 Yue-Yang Road, Shanghai 200031, China.
Organizational Affiliation: