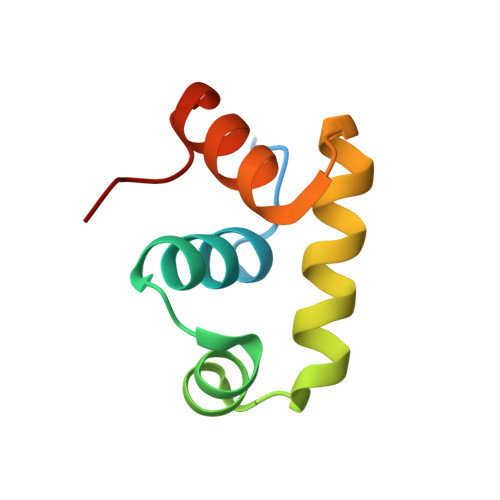
Crystal structure of a transcription factor, GerE (PaGerE), from spore-forming bacterium Paenisporosarcina sp. TG-14.
Lee, C.W., Park, S.H., Koh, H.Y., Jeong, C.S., Hwang, J., Lee, S.G., Youn, U.J., Lee, C.S., Park, H.H., Kim, H.J., Park, H., Lee, J.H.(2019) Biochem Biophys Res Commun 513: 374-379
- PubMed: 30967265
- DOI: https://doi.org/10.1016/j.bbrc.2019.04.019
- Primary Citation of Related Structures:
6JQS - PubMed Abstract:
In cold and harsh environments such as glaciers and sediments in ice cores, microbes can survive by forming spores. Spores are composed of a thick coat protein, which protects against external factors such as heat-shock, high salinity, and nutrient deficiency. GerE is a key transcription factor involved in spore coat protein expression in the mother cell during sporulation. GerE regulates transcription during the late sporulation stage by directly binding to the promoter of cotB gene. Here, we report the crystal structure of PaGerE at 2.09 Å resolution from Paenisporosarcina sp. TG-14, which was isolated from the Taylor glacier. The PaGerE structure is composed of four α-helices and adopts a helix-turn-helix architecture with 68 amino acid residues. Based on our DNA binding analysis, the PaGerE binds to the promoter region of CotB to affect protein expression. Additionally, our structural comparison studies suggest that DNA binding by PaGerE causes a conformational change in the α4-helix region, which may strongly induce dimerization of PaGerE.
- Unit of Polar Genomics, Korea Polar Research Institute, Incheon, 21990, Republic of Korea.
Organizational Affiliation: