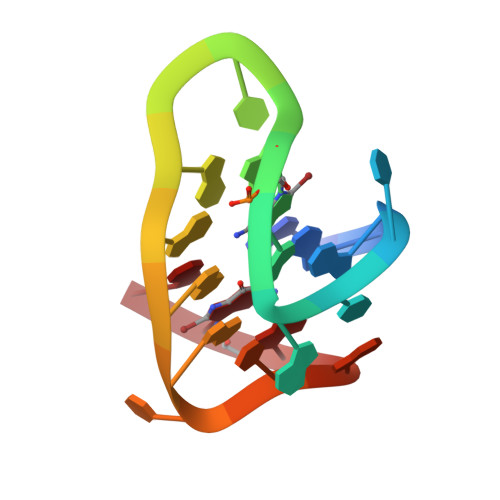
The crystal structure of an antiparallel chair-type G-quadruplex formed by Bromo-substituted human telomeric DNA.
Geng, Y., Liu, C., Zhou, B., Cai, Q., Miao, H., Shi, X., Xu, N., You, Y., Fung, C.P., Din, R.U., Zhu, G.(2019) Nucleic Acids Res 47: 5395-5404
- PubMed: 30957851
- DOI: https://doi.org/10.1093/nar/gkz221
- Primary Citation of Related Structures:
6JKN - PubMed Abstract:
Human telomeric guanine-rich DNA, which could adopt different G-quadruplex structures, plays important roles in protecting the cell from recombination and degradation. Although many of these structures were determined, the chair-type G-quadruplex structure remains elusive. Here, we present a crystal structure of the G-quadruplex composed of the human telomeric sequence d[GGGTTAGG8GTTAGGGTTAGG20G] with two dG to 8Br-dG substitutions at positions 8 and 20 with syn conformation in the K+ solution. It forms a novel three-layer chair-type G-quadruplex with two linking trinucleotide loops. Particularly, T5 and T17 are coplanar with two water molecules stacking on the G-tetrad layer in a sandwich-like mode through a coordinating K+ ion and an A6•A18 base pair. While a twisted Hoogsteen A12•T10 base pair caps on the top of G-tetrad core. The three linking TTA loops are edgewise and each DNA strand has two antiparallel adjacent strands. Our findings contribute to a deeper understanding and highlight the unique roles of loop and water molecule in the folding of the G-quadruplex.
- Division of Life Science, The Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong SAR, China.
Organizational Affiliation: