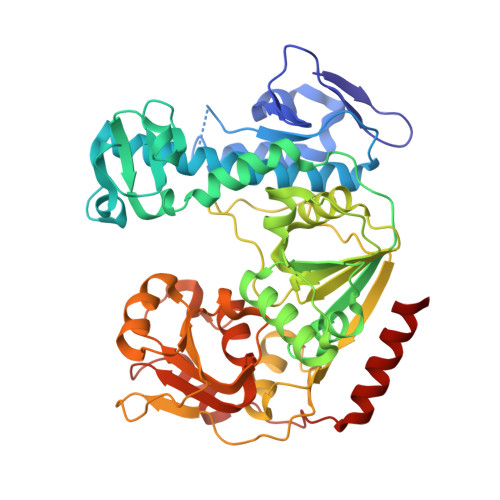
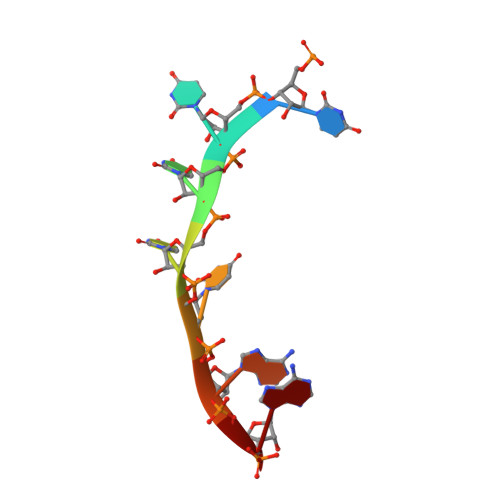
Structural insights into RNA recognition by the Chikungunya virus nsP2 helicase.
Law, Y.S., Utt, A., Tan, Y.B., Zheng, J., Wang, S., Chen, M.W., Griffin, P.R., Merits, A., Luo, D.(2019) Proc Natl Acad Sci U S A 116: 9558-9567
- PubMed: 31000599
- DOI: https://doi.org/10.1073/pnas.1900656116
- Primary Citation of Related Structures:
6JIM - PubMed Abstract:
Chikungunya virus (CHIKV) is transmitted to humans through mosquitoes and causes Chikungunya fever. Nonstructural protein 2 (nsP2) exhibits the protease and RNA helicase activities that are required for viral RNA replication and transcription. Unlike for the C-terminal protease, the structure of the N-terminal RNA helicase (nsP2h) has not been determined. Here, we report the crystal structure of the nsP2h bound to the conserved 3'-end 14 nucleotides of the CHIKV genome and the nonhydrolyzable transition-state nucleotide analog ADP-AlF 4 Overall, the structural analysis revealed that nsP2h adopts a uniquely folded N-terminal domain followed by a superfamily 1 RNA helicase fold. The conserved helicase motifs establish polar contacts with the RNA backbone. There are three hydrophobic residues (Y161, F164, and F287) which form stacking interactions with RNA bases and thereby bend the RNA backbone. An F287A substitution that disrupted these stacking interactions increased the basal ATPase activity but decreased the RNA binding affinity. Furthermore, the F287A substitution reduced viral infectivity by attenuating subgenomic RNA synthesis. Replication of the mutant virus was restored by pseudoreversion (A287V) or adaptive mutations in the RecA2 helicase domain (T358S or V410I). Y161A and/or F164A substitutions, which were designed to disrupt the interactions with the RNA molecule, did not affect the ATPase activity but completely abolished the replication and transcription of viral RNA and the infectivity of CHIKV. Our study sheds light on the roles of the RNA helicase region in viral replication and provides insights that might be applicable to alphaviruses and other RNA viruses in general.
- Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore 636921.
Organizational Affiliation: