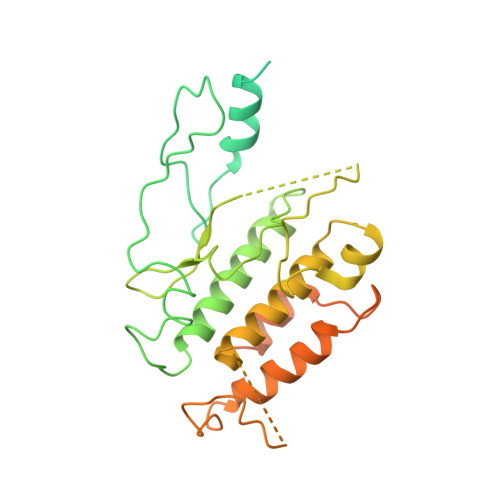
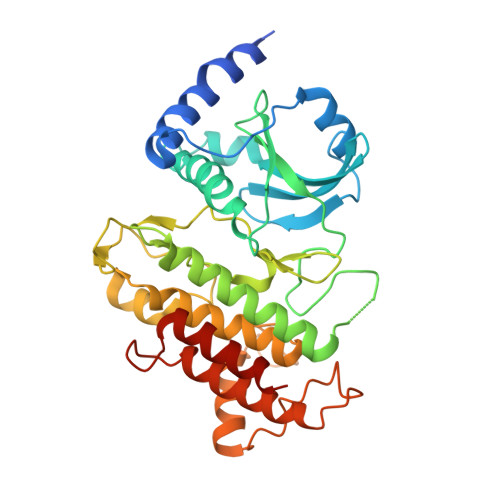
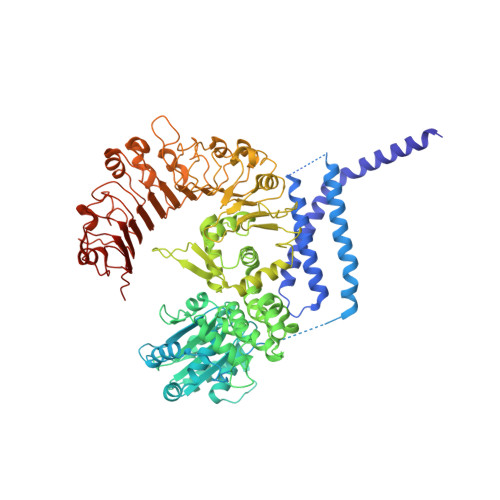
Reconstitution and structure of a plant NLR resistosome conferring immunity.
Wang, J., Hu, M., Wang, J., Qi, J., Han, Z., Wang, G., Qi, Y., Wang, H.W., Zhou, J.M., Chai, J.(2019) Science 364
- PubMed: 30948527
- DOI: https://doi.org/10.1126/science.aav5870
- Primary Citation of Related Structures:
6J5T, 6J6I - PubMed Abstract:
Nucleotide-binding, leucine-rich repeat receptors (NLRs) perceive pathogen effectors to trigger plant immunity. Biochemical mechanisms underlying plant NLR activation have until now remained poorly understood. We reconstituted an active complex containing the Arabidopsis coiled-coil NLR ZAR1, the pseudokinase RKS1, uridylated protein kinase PBL2, and 2'-deoxyadenosine 5'-triphosphate (dATP), demonstrating the oligomerization of the complex during immune activation. The cryo-electron microscopy structure reveals a wheel-like pentameric ZAR1 resistosome. Besides the nucleotide-binding domain, the coiled-coil domain of ZAR1 also contributes to resistosome pentamerization by forming an α-helical barrel that interacts with the leucine-rich repeat and winged-helix domains. Structural remodeling and fold switching during activation release the very N-terminal amphipathic α helix of ZAR1 to form a funnel-shaped structure that is required for the plasma membrane association, cell death triggering, and disease resistance, offering clues to the biochemical function of a plant resistosome.
- Beijing Advanced Innovation Center for Structural Biology, Tsinghua-Peking Joint Center for Life Sciences, Center for Plant Biology, School of Life Sciences, Tsinghua University, 100084 Beijing, China.
Organizational Affiliation: