Structural basis for the Target DNA recognition and binding by the MYB domain of phosphate starvation response 1.
Jiang, M., Sun, L., Isupov, M.N., Littlechild, J.A., Wu, X., Wang, Q., Wang, Q., Yang, W., Wu, Y.(2019) FEBS J 286: 2809-2821
- PubMed: 30974511
- DOI: https://doi.org/10.1111/febs.14846
- Primary Citation of Related Structures:
6J4K, 6J4R, 6J5B - PubMed Abstract:
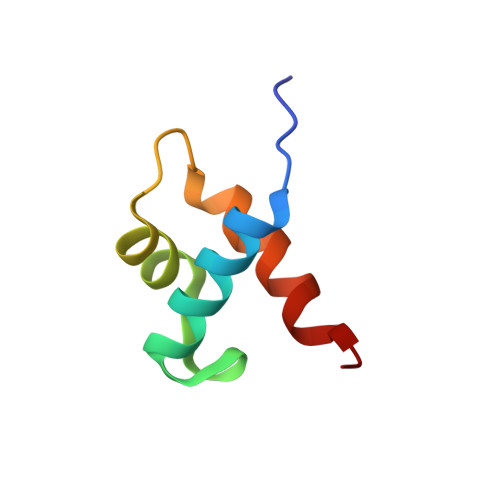
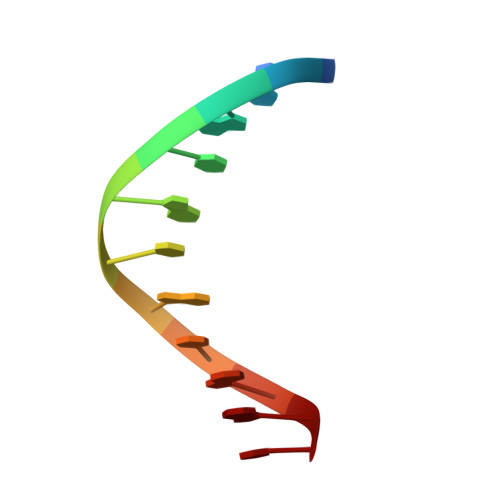
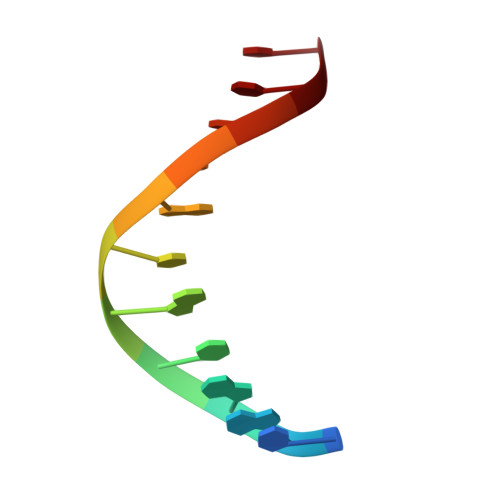
The phosphate starvation response 1 (PHR1) protein has a central role in mediating the response to phosphate starvation in plants. PHR1 is composed of a number of domains including a MYB domain involved with DNA binding and a coiled-coil domain proposed to be involved with dimer formation. PHR1 binds to the promoter of phosphate starvation-induced genes to control the levels of phosphate required for nutrition. Previous studies have shown that both the MYB domain and the coiled-coil domain of PHR1 are required for binding the target DNA. Here, we describe the crystal structure of the PHR1 MYB domain and two structures of its complex with the PHR1-binding DNA sequence (P1BS). Structural and isothermal titration calorimetry has been carried out showing that the MYB domain of PHR1 alone is sufficient for target DNA recognition and binding. Two copies of the PHR1 MYB domain bind to the same major groove of the P1BS DNA with few direct interactions between the individual MYB domains. In addition, the PHR1 MYB-P1BS DNA complex structures reveal amino acid residues involved in DNA recognition and binding. Mutagenesis of these residues results in lost or impaired ability of PHR1 MYB to bind to its target DNA. The results presented reveal the structural basis for DNA recognition by the PHR1 MYB domain and demonstrate that two PHR1 MYB domains attach to their P1BS DNA targeting sequence. DATABASE: Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 6J4K (PHR1 MYB), 6J4R (PHR1 MYB-R-P1BS), 6J5B (MYB-CC-R2-P1BS).
- State Key Laboratory of Structural Chemistry, Fujian Institute of Research on the Structure of Matter, Chinese Academy of Sciences, Fuzhou, China.
Organizational Affiliation: