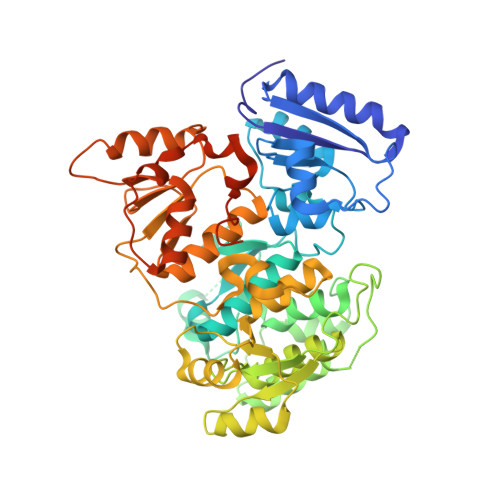
Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Wan, B., Wu, J., Meng, X., Lei, M., Zhao, X.(2019) Mol Cell 75: 238-251.e5
- PubMed: 31348879
- DOI: https://doi.org/10.1016/j.molcel.2019.05.035
- Primary Citation of Related Structures:
6J0V, 6J0W, 6J0X, 6J0Y - PubMed Abstract:
BRCT domains support myriad protein-protein interactions involved in genome maintenance. Although di-BRCT recognition of phospho-proteins is well known to support the genotoxic response, whether multi-BRCT domains can acquire distinct structures and functions is unclear. Here we present the tetra-BRCT structures from the conserved yeast protein Rtt107 in free and ligand-bound forms. The four BRCT repeats fold into a tetrahedral structure that recognizes unmodified ligands using a bi-partite mechanism, suggesting repeat origami enabling function acquisition. Functional studies show that Rtt107 binding of partner proteins of diverse activities promotes genome replication and stability in both distinct and concerted manners. A unified theme is that tetra- and di-BRCT domains of Rtt107 collaborate to recruit partner proteins to chromatin. Our work thus illustrates how a master regulator uses two types of BRCT domains to recognize distinct genome factors and direct them to chromatin for constitutive genome protection.
- Molecular Biology Department, Memorial Sloan Kettering Cancer Center, New York, NY 10065, USA.
Organizational Affiliation: