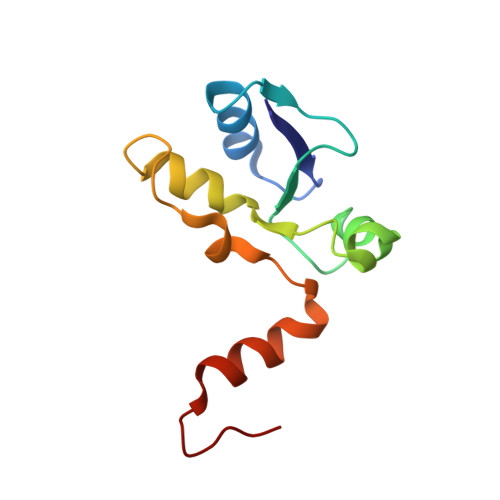
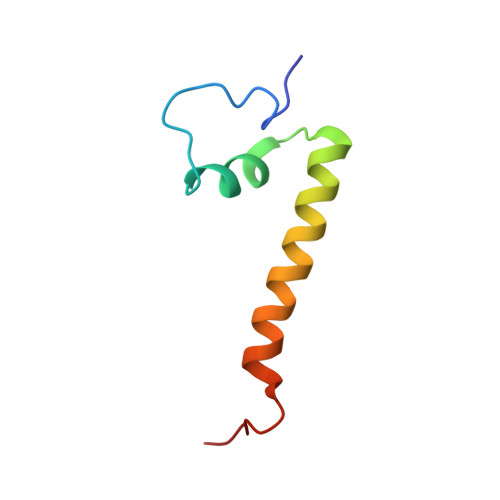
The meiotic TERB1-TERB2-MAJIN complex tethers telomeres to the nuclear envelope.
Wang, Y., Chen, Y., Chen, J., Wang, L., Nie, L., Long, J., Chang, H., Wu, J., Huang, C., Lei, M.(2019) Nat Commun 10: 564-564
- PubMed: 30718482
- DOI: https://doi.org/10.1038/s41467-019-08437-1
- Primary Citation of Related Structures:
6J07, 6J08 - PubMed Abstract:
During meiotic prophase I, telomeres attach to and move on the nuclear envelope (NE), regulating chromosome movement to promote homologous pairing. Meiosis-specific proteins TERB1, TERB2 and MAJIN play a key role in this process. Here, we report the crystal structures of human TERB1-TERB2 and TERB2-MAJIN subcomplexes. Specific disruption of the TERB1-TERB2 or the TERB2-MAJIN interaction in the mouse Terb2 gene abolishes the telomere attachment to the NE and causes aberrant homologous pairing and disordered synapsis. In addition, depletion of SUN1 also partially disrupts the telomere-NE connection. We propose that the telomere-TRF1-TERB1-TERB2-MAJIN-NE interaction network and the telomere-LINC complex connection are likely two separate but cooperative pathways to stably recruit telomeres to the NE in meiosis prophase I. Our work provides a molecular model of the connection between telomeres and the NE and reveals the correlation between aberrant synapsis and the defective telomere attachment to the NE.
- National Center for Protein Science Shanghai, State Key Laboratory of Molecular Biology, CAS Center for Excellence on Molecular Cell Science, Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, University of Chinese Academy of Sciences, 200031, Shanghai, China.
Organizational Affiliation: