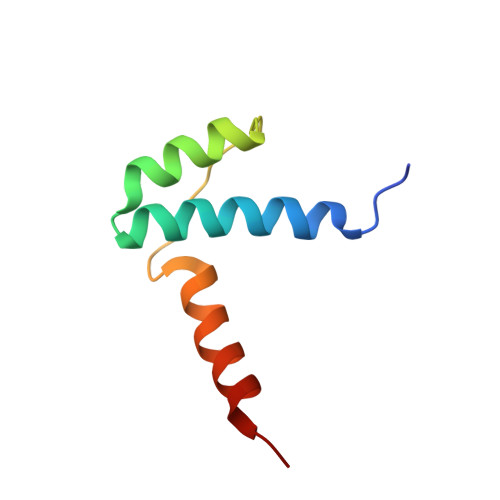
Crystal structure of the LUFS domain of human single-stranded DNA binding Protein 2 (SSBP2).
Wang, H., Wang, Z., Tang, Q., Yan, X.X., Xu, W.(2019) Protein Sci 28: 788-793
- PubMed: 30676665
- DOI: https://doi.org/10.1002/pro.3581
- Primary Citation of Related Structures:
6IWV - PubMed Abstract:
The human single-stranded DNA binding Protein 2 (SSBP2) is a tumor suppressor implicated in multiple cancer forms. The SSBP2 and related SSBP3/SSBP4 proteins are predicted to be intrinsically disordered excepted for their highly conserved N-terminal LUFS (LUG/LUH, Flo8, and SSBP/SSDP) domain. LUFS domains are found in a number of proteins including some transcriptional co-repressors. Although LUFS domains contain an N-terminal Lis homology (LisH) motif that typically forms a stable dimer, no 3D structure of any LUFS domain is available. Here, we report a crystal structure of the LUFS domain of human SSBP2 at 1.52 Å resolution. We show that the SSBP2 LUFS domain forms a homo-tetramer and reveal how an alpha-helix C-terminal to the LisH motif mediates SSBP2 tetramerization (dimerization of dimers). Conservation of the tetramerization interface among LUFS domains suggests that other LUFS domains may also form tetramers in similar manners.
- National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, People's Republic of China.
Organizational Affiliation: