structural insights into Mycobacterium tuberculosis ClpP1P2 inhibition by Cediranib: implications for developing antimicrobial agents targeting Clp protease
Bao, R., Luo, Y.F., Zhu, Y.B., Yang, Y., Yang, T.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP-dependent Clp protease proteolytic subunit 2 | 202 | Mycobacterium tuberculosis CDC1551 | Mutation(s): 0 Gene Names: clpP2, MT2535 EC: 3.4.21.92 | 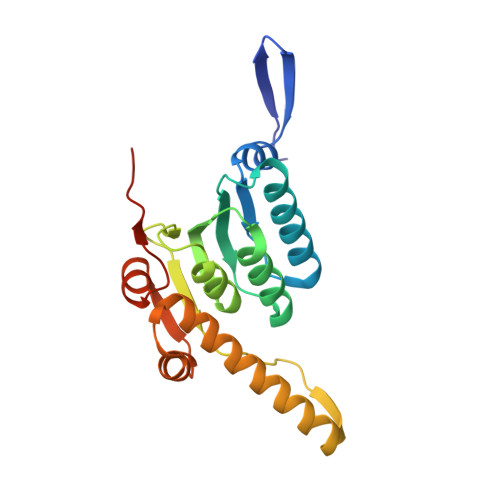 | |
UniProt | |||||
Find proteins for P9WPC3 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WPC3 Go to UniProtKB: P9WPC3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WPC3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP-dependent Clp protease proteolytic subunit 1 | 194 | Mycobacterium tuberculosis CDC1551 | Mutation(s): 0 Gene Names: clpP1, clpP, MT2536 EC: 3.4.21.92 | 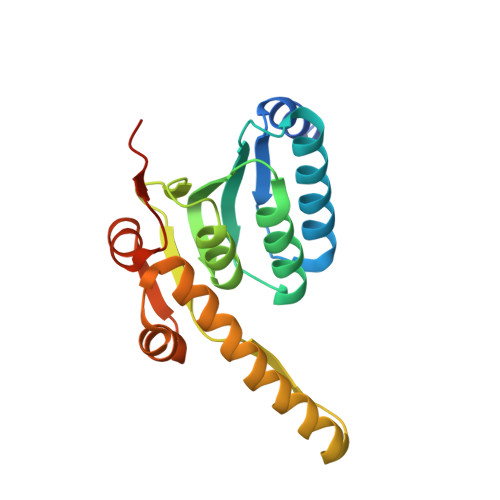 | |
UniProt | |||||
Find proteins for P9WPC5 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WPC5 Go to UniProtKB: P9WPC5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WPC5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| AV3 (Subject of Investigation/LOI) Query on AV3 | DA [auth I] IA [auth K] PA [auth N] Q [auth B] R [auth B] | 4-[(4-fluoro-2-methyl-3H-indol-5-yl)oxy]-6-methoxy-7-[3-(pyrrolidin-1-yl)propoxy]quinazoline C25 H27 F N4 O3 ZOVVOYNGCNKFNN-UHFFFAOYSA-N |  | ||
| S0R Query on S0R | BA [auth H] EA [auth I] GA [auth J] JA [auth K] LA [auth L] | (2S)-2-benzamido-4-methyl-pentanoic acid C13 H17 N O3 POLGZPYHEPOBFG-NSHDSACASA-N |  | ||
| LEU Query on LEU | AA [auth F] CA [auth H] FA [auth I] HA [auth J] KA [auth K] | LEUCINE C6 H13 N O2 ROHFNLRQFUQHCH-YFKPBYRVSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 210.634 | α = 90 |
| b = 180.78 | β = 95.716 |
| c = 95.926 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 81473253 |