Discovery and characterisation of an antibody that selectively modulates the inhibitory activity of plasminogen activator inhibitor-1.
Vousden, K.A., Lundqvist, T., Popovic, B., Naiman, B., Carruthers, A.M., Newton, P., Johnson, D.J.D., Pomowski, A., Wilkinson, T., Dufner, P., de Mendez, I., Mallinder, P.R., Murray, C., Strain, M., Connor, J., Murray, L.A., Sleeman, M.A., Lowe, D.C., Huntington, J.A., Vaughan, T.J.(2019) Sci Rep 9: 1605-1605
- PubMed: 30733557
- DOI: https://doi.org/10.1038/s41598-019-38842-x
- Primary Citation of Related Structures:
6I8S - PubMed Abstract:
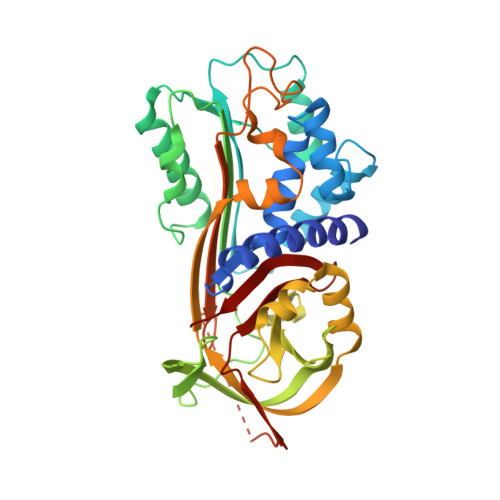
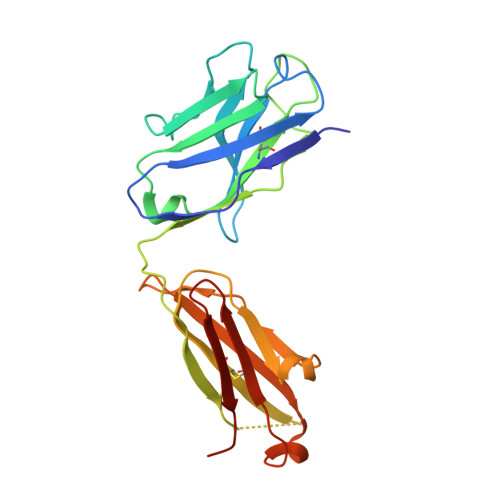
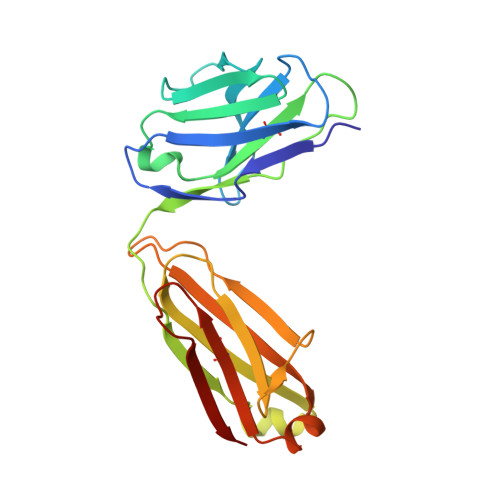
Plasminogen activator inhibitor-1 (PAI-1) is a serine protease inhibitor (serpin) that regulates fibrinolysis, cell adhesion and cell motility via its interactions with plasminogen activators and vitronectin. PAI-1 has been shown to play a role in a number of diverse pathologies including cardiovascular diseases, obesity and cancer and is therefore an attractive therapeutic target. However the multiple patho-physiological roles of PAI-1, and understanding the relative contributions of these in any one disease setting, make the development of therapeutically relevant molecules challenging. Here we describe the identification and characterisation of fully human antibody MEDI-579, which binds with high affinity and specificity to the active form of human PAI-1. MEDI-579 specifically inhibits serine protease interactions with PAI-1 while conserving vitronectin binding. Crystallographic analysis reveals that this specificity is achieved through direct binding of MEDI-579 Fab to the reactive centre loop (RCL) of PAI-1 and at the same exosite used by both tissue and urokinase plasminogen activators (tPA and uPA). We propose that MEDI-579 acts by directly competing with proteases for RCL binding and as such is able to modulate the interaction of PAI-1 with tPA and uPA in a way not previously described for a human PAI-1 inhibitor.
- MedImmune Ltd, Cambridge, CB21 6GH, UK. vousdenk@medimmune.com.
Organizational Affiliation: