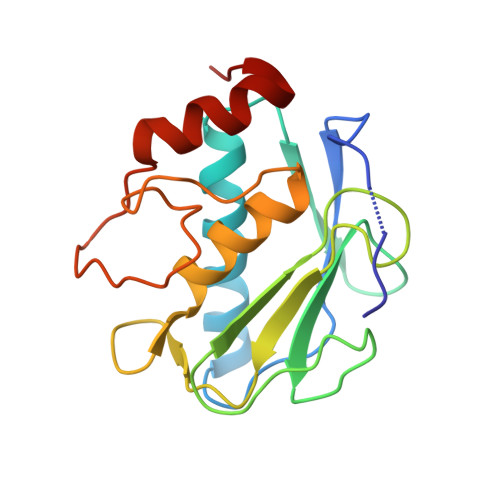
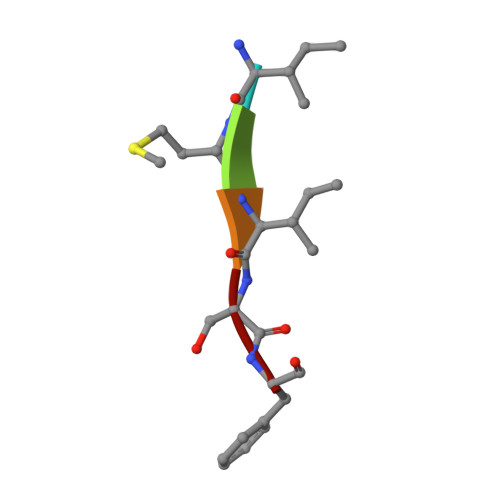
Drug Design Inspired by Nature: Crystallographic Detection of an Auto-Tailored Protease Inhibitor Template.
Gall, F.M., Hohl, D., Frasson, D., Wermelinger, T., Mittl, P.R.E., Sievers, M., Riedl, R.(2019) Angew Chem Int Ed Engl 58: 4051-4055
- PubMed: 30615822
- DOI: https://doi.org/10.1002/anie.201812348
- Primary Citation of Related Structures:
6HV2 - PubMed Abstract:
De novo drug discovery is still a challenge in the search for potent and selective modulators of therapeutically relevant target proteins. Here, we disclose the unexpected discovery of a peptidic ligand 1 by X-ray crystallography, which was auto-tailored by the therapeutic target MMP-13 through partial self-degradation and subsequent structure-based optimization to a highly potent and selective β-sheet peptidomimetic inhibitor derived from the endogenous tissue inhibitors of metalloproteinases (TIMPs). The incorporation of non-proteinogenic amino acids in combination with a cyclization strategy proved to be key for the de novo design of TIMP peptidomimetics. The optimized cyclic peptide 4 (ZHAWOC7726) is membrane permeable with an IC 50 of 21 nm for MMP-13 and an attractive selectivity profile with respect to a polypharmacology approach including the anticancer targets MMP-2 (IC 50 : 170 nm) and MMP-9 (IC 50 : 140 nm).
- Institute of Chemistry and Biotechnology, Center of Organic and Medicinal Chemistry, ZHAW Zurich University of Applied Sciences, Einsiedlerstrasse 31, 8820, Wädenswil, Switzerland.
Organizational Affiliation: