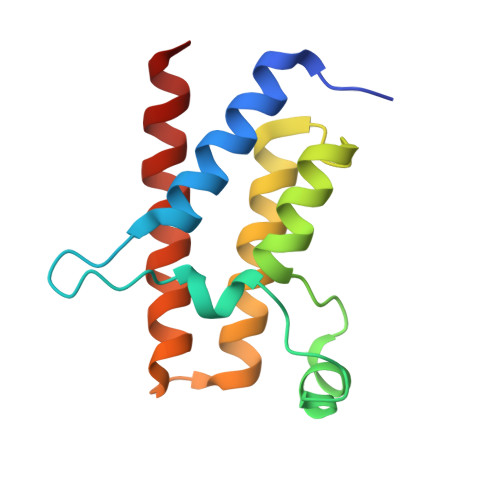
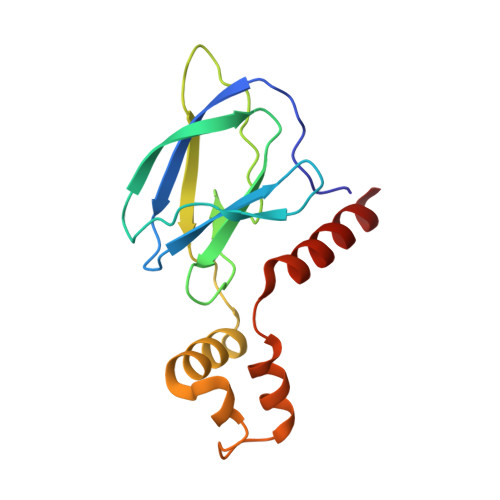
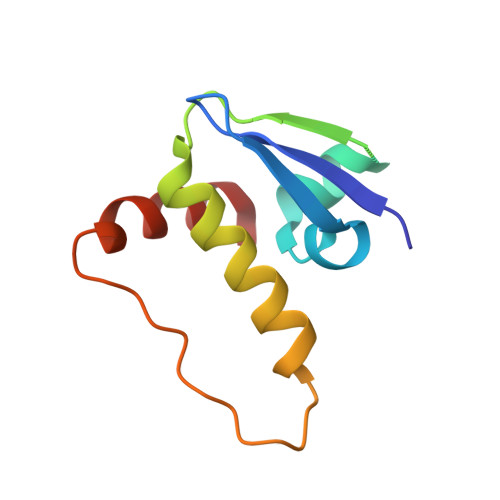
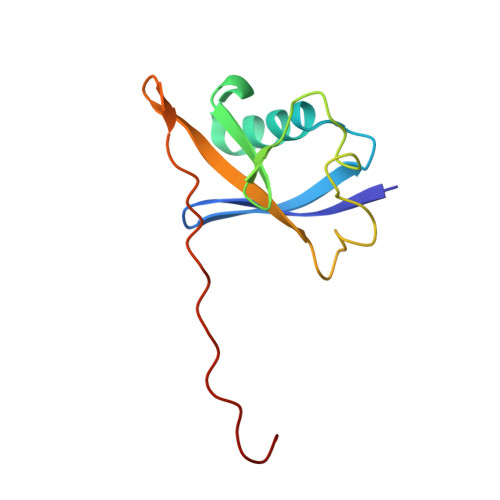
BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design.
Farnaby, W., Koegl, M., Roy, M.J., Whitworth, C., Diers, E., Trainor, N., Zollman, D., Steurer, S., Karolyi-Oezguer, J., Riedmueller, C., Gmaschitz, T., Wachter, J., Dank, C., Galant, M., Sharps, B., Rumpel, K., Traxler, E., Gerstberger, T., Schnitzer, R., Petermann, O., Greb, P., Weinstabl, H., Bader, G., Zoephel, A., Weiss-Puxbaum, A., Ehrenhofer-Wolfer, K., Wohrle, S., Boehmelt, G., Rinnenthal, J., Arnhof, H., Wiechens, N., Wu, M.Y., Owen-Hughes, T., Ettmayer, P., Pearson, M., McConnell, D.B., Ciulli, A.(2019) Nat Chem Biol 15: 672-680
- PubMed: 31178587
- DOI: https://doi.org/10.1038/s41589-019-0294-6
- Primary Citation of Related Structures:
6HAX, 6HAY, 6HAZ, 6HR2 - PubMed Abstract:
Targeting subunits of BAF/PBAF chromatin remodeling complexes has been proposed as an approach to exploit cancer vulnerabilities. Here, we develop proteolysis targeting chimera (PROTAC) degraders of the BAF ATPase subunits SMARCA2 and SMARCA4 using a bromodomain ligand and recruitment of the E3 ubiquitin ligase VHL. High-resolution ternary complex crystal structures and biophysical investigation guided rational and efficient optimization toward ACBI1, a potent and cooperative degrader of SMARCA2, SMARCA4 and PBRM1. ACBI1 induced anti-proliferative effects and cell death caused by SMARCA2 depletion in SMARCA4 mutant cancer cells, and in acute myeloid leukemia cells dependent on SMARCA4 ATPase activity. These findings exemplify a successful biophysics- and structure-based PROTAC design approach to degrade high profile drug targets, and pave the way toward new therapeutics for the treatment of tumors sensitive to the loss of BAF complex ATPases.
- Division of Biological Chemistry and Drug Discovery, School of Life Sciences, University of Dundee, Dundee, UK.
Organizational Affiliation: