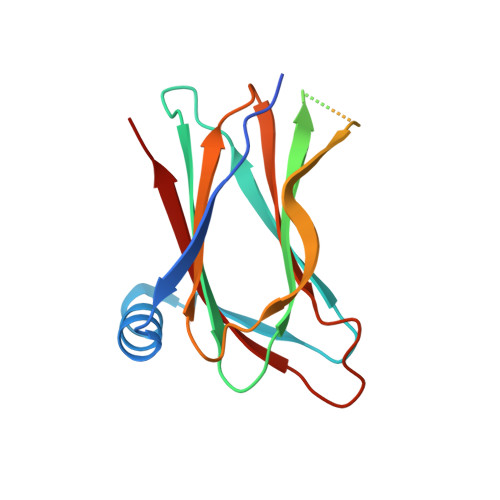
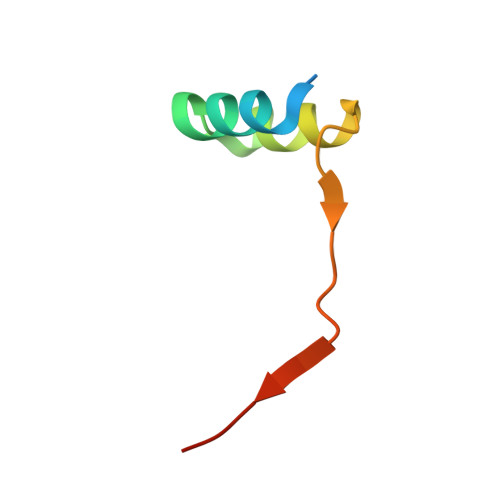
Convergent evolution in the mechanisms of ACBD3 recruitment to picornavirus replication sites.
Horova, V., Lyoo, H., Rozycki, B., Chalupska, D., Smola, M., Humpolickova, J., Strating, J.R.P.M., van Kuppeveld, F.J.M., Boura, E., Klima, M.(2019) PLoS Pathog 15: e1007962-e1007962
- PubMed: 31381608
- DOI: https://doi.org/10.1371/journal.ppat.1007962
- Primary Citation of Related Structures:
6HLN, 6HLT, 6HLV, 6HLW, 6HM8, 6HMV - PubMed Abstract:
Enteroviruses, members of the family of picornaviruses, are the most common viral infectious agents in humans causing a broad spectrum of diseases ranging from mild respiratory illnesses to life-threatening infections. To efficiently replicate within the host cell, enteroviruses hijack several host factors, such as ACBD3. ACBD3 facilitates replication of various enterovirus species, however, structural determinants of ACBD3 recruitment to the viral replication sites are poorly understood. Here, we present a structural characterization of the interaction between ACBD3 and the non-structural 3A proteins of four representative enteroviruses (poliovirus, enterovirus A71, enterovirus D68, and rhinovirus B14). In addition, we describe the details of the 3A-3A interaction causing the assembly of the ACBD3-3A heterotetramers and the interaction between the ACBD3-3A complex and the lipid bilayer. Using structure-guided identification of the point mutations disrupting these interactions, we demonstrate their roles in the intracellular localization of these proteins, recruitment of downstream effectors of ACBD3, and facilitation of enterovirus replication. These structures uncovered a striking convergence in the mechanisms of how enteroviruses and kobuviruses, members of a distinct group of picornaviruses that also rely on ACBD3, recruit ACBD3 and its downstream effectors to the sites of viral replication.
- Institute of Organic Chemistry and Biochemistry, Czech Academy of Sciences, Prague, Czech Republic.
Organizational Affiliation: