Structure-Guided Exploration of SDS22 Interactions with Protein Phosphatase PP1 and the Splicing Factor BCLAF1.
Heroes, E., Van der Hoeven, G., Choy, M.S., Garcia, J.D.P., Ferreira, M., Nys, M., Derua, R., Beullens, M., Ulens, C., Peti, W., Van Meervelt, L., Page, R., Bollen, M.(2019) Structure 27: 507-518.e5
- PubMed: 30661852
- DOI: https://doi.org/10.1016/j.str.2018.12.002
- Primary Citation of Related Structures:
6HKW - PubMed Abstract:
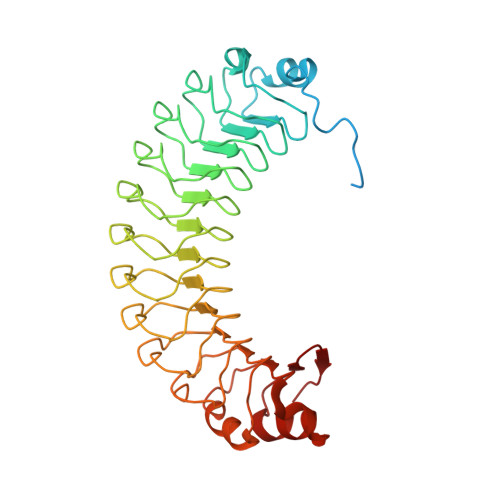
SDS22 is an ancient regulator of protein phosphatase-1 (PP1). Our crystal structure of SDS22 shows that its twelve leucine-rich repeats adopt a banana-shaped fold that is shielded from solvent by capping domains at its extremities. Subsequent modeling and biochemical studies revealed that the concave side of SDS22 likely interacts with PP1 helices α5 and α6, which are distal from the binding sites of many previously described PP1 interactors. Accordingly, we found that SDS22 acts as a "third" subunit of multiple PP1 holoenzymes. The crystal structure of SDS22 also revealed a large basic surface patch that enables binding of a phosphorylated form of splicing factor BCLAF1. Taken together, our data provide insights into the formation of PP1:SDS22 and the recruitment of additional interaction proteins, such as BCLAF1.
- Laboratory of Biosignaling & Therapeutics, Department of Cellular and Molecular Medicine, KU Leuven, Leuven, Belgium; Biomolecular Architecture, Department of Chemistry, KU Leuven, Leuven, Belgium.
Organizational Affiliation: