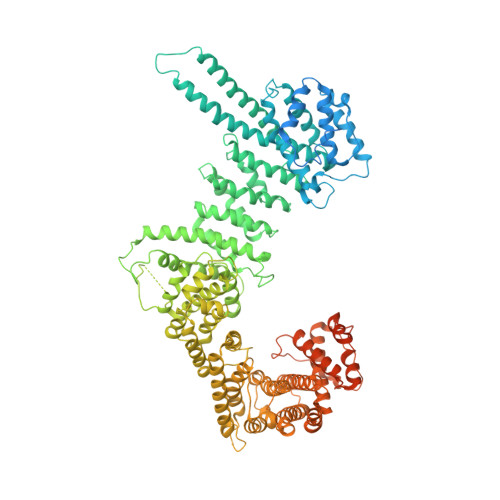
Structural basis for Scc3-dependent cohesin recruitment to chromatin.
Li, Y., Muir, K., Bowler, M.W., Metz, J., Haering, C.H., Panne, D.(2018) Elife 7
- PubMed: 30109982
- DOI: https://doi.org/10.7554/eLife.38356
- Primary Citation of Related Structures:
6H8Q - PubMed Abstract:
The cohesin ring complex is required for numerous chromosomal transactions including sister chromatid cohesion, DNA damage repair and transcriptional regulation. How cohesin engages its chromatin substrate has remained an unresolved question. We show here, by determining a crystal structure of the budding yeast cohesin HEAT-repeat subunit Scc3 bound to a fragment of the Scc1 kleisin subunit and DNA, that Scc3 and Scc1 form a composite DNA interaction module. The Scc3-Scc1 subcomplex engages double-stranded DNA through a conserved, positively charged surface. We demonstrate that this conserved domain is required for DNA binding by Scc3-Scc1 in vitro, as well as for the enrichment of cohesin on chromosomes and for cell viability. These findings suggest that the Scc3-Scc1 DNA-binding interface plays a central role in the recruitment of cohesin complexes to chromosomes and therefore for cohesin to faithfully execute its functions during cell division.
- European Molecular Biology Laboratory, Grenoble, France.
Organizational Affiliation: