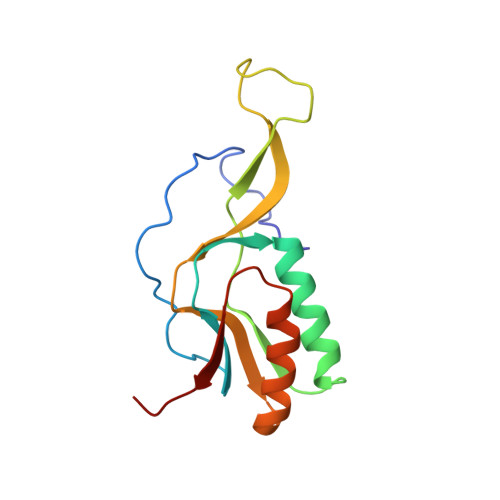
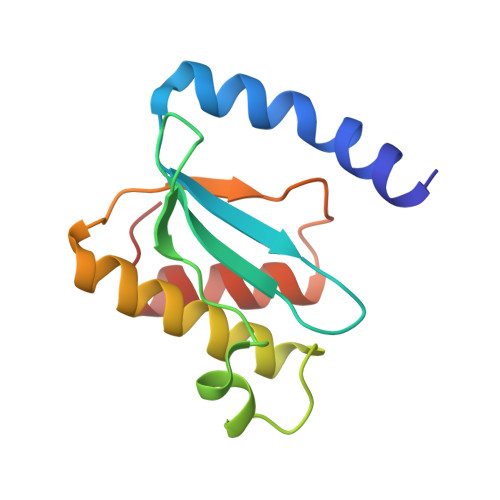
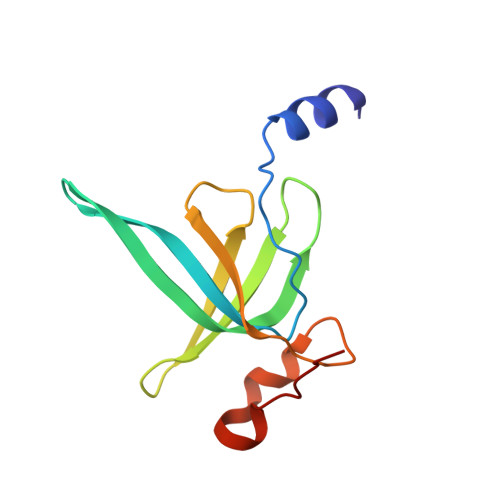
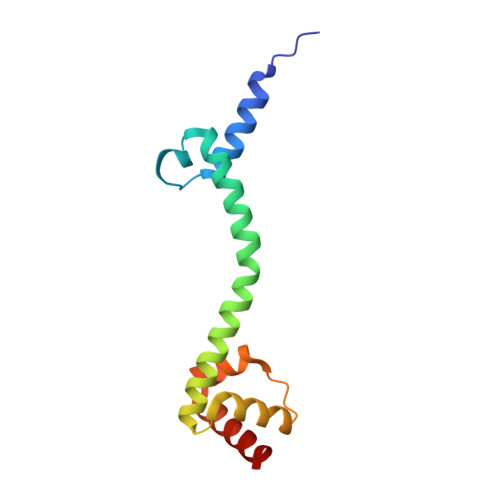
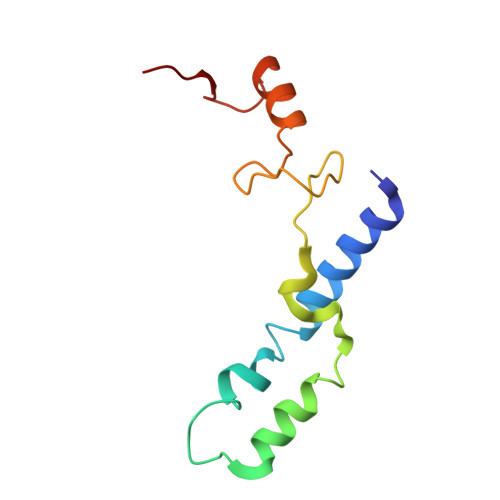
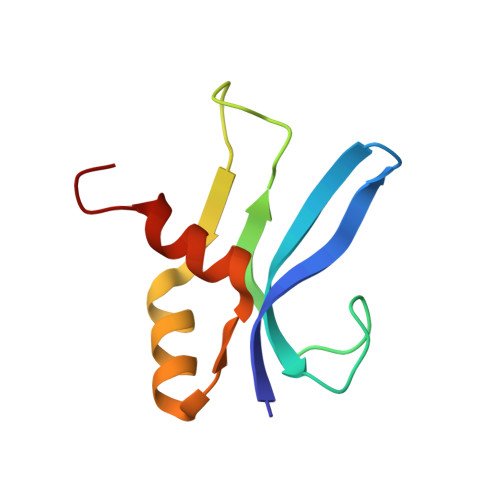
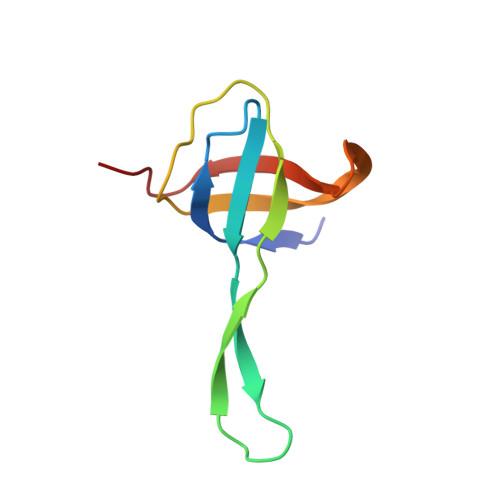
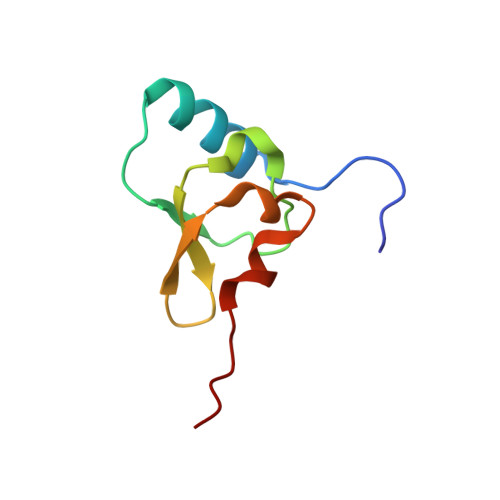
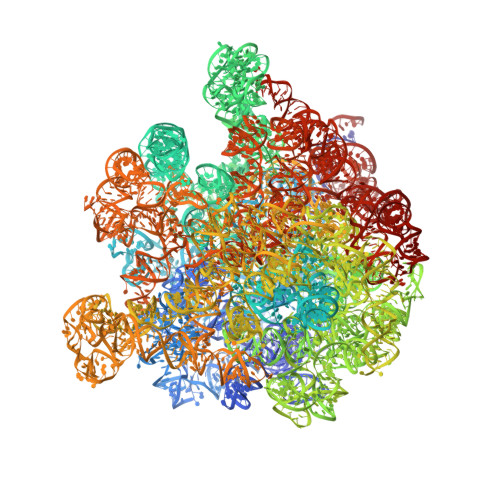
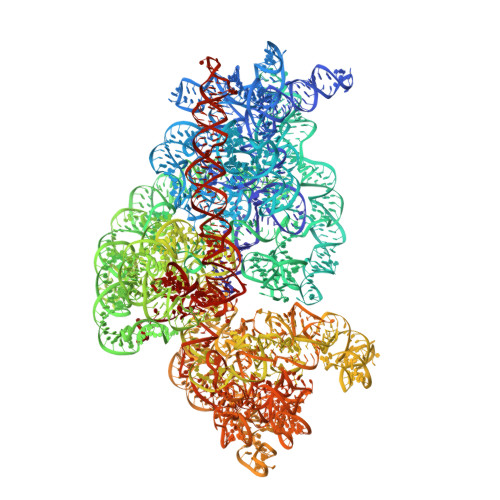
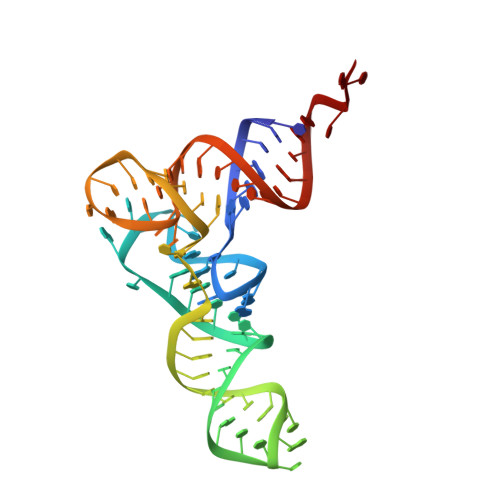
Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1.
Beckert, B., Turk, M., Czech, A., Berninghausen, O., Beckmann, R., Ignatova, Z., Plitzko, J.M., Wilson, D.N.(2018) Nat Microbiol 3: 1115-1121
- PubMed: 30177741
- DOI: https://doi.org/10.1038/s41564-018-0237-0
- Primary Citation of Related Structures:
6H4N, 6H58 - PubMed Abstract:
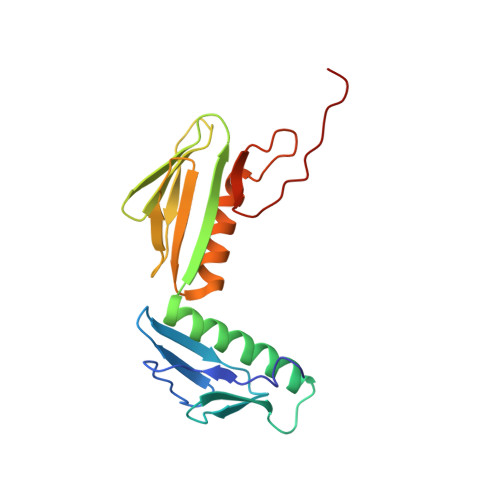
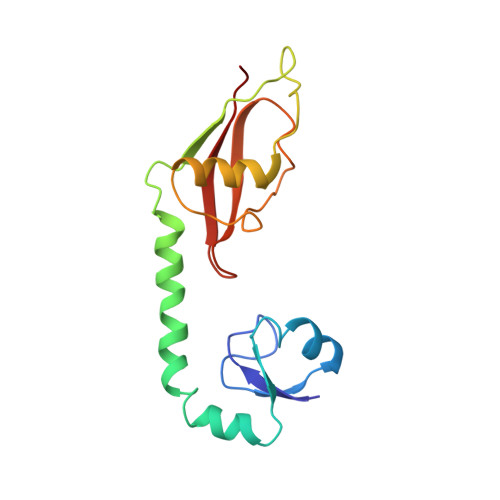
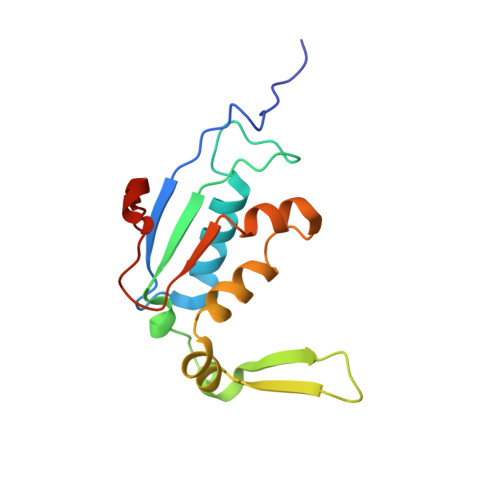
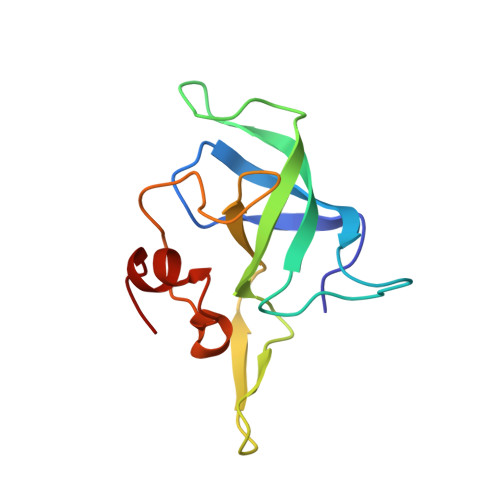
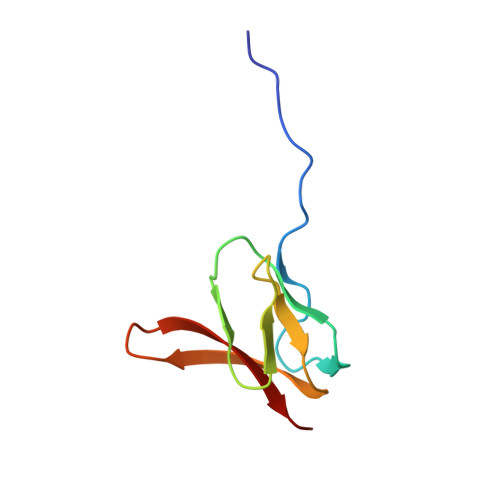
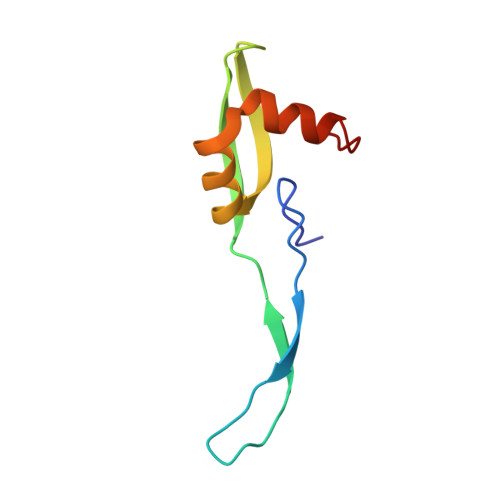
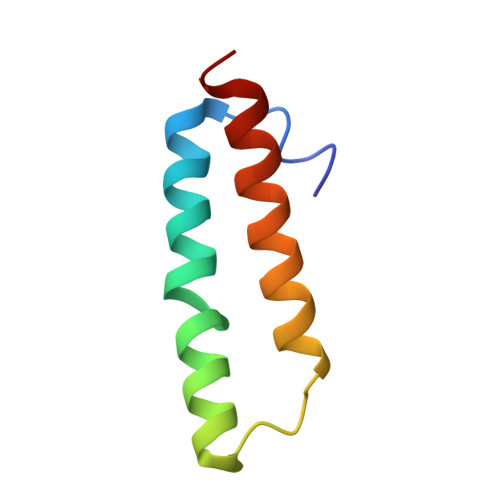
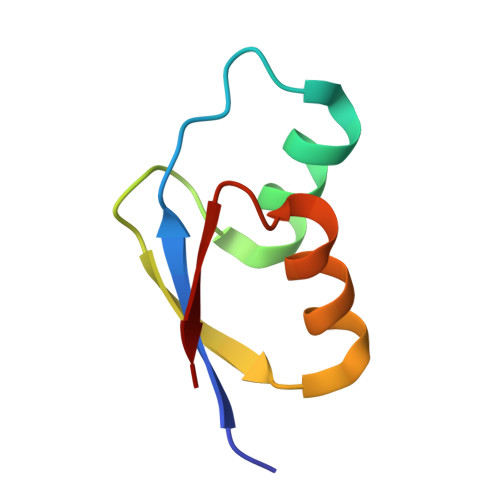
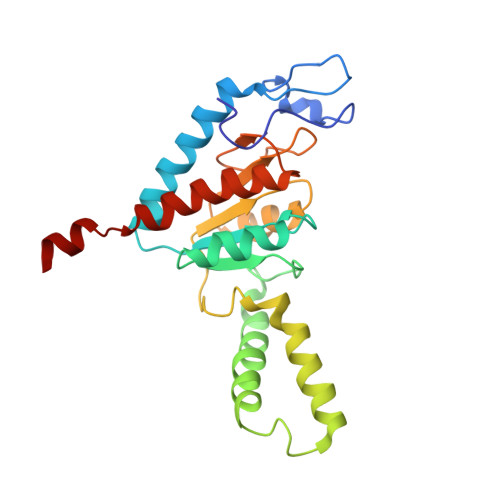
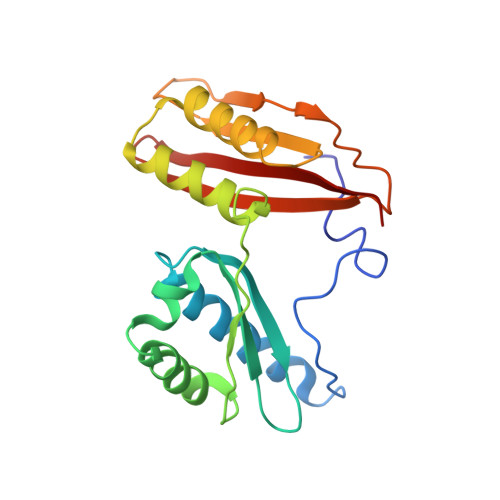
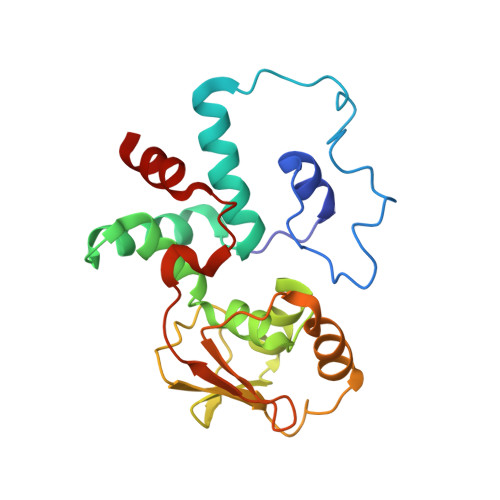
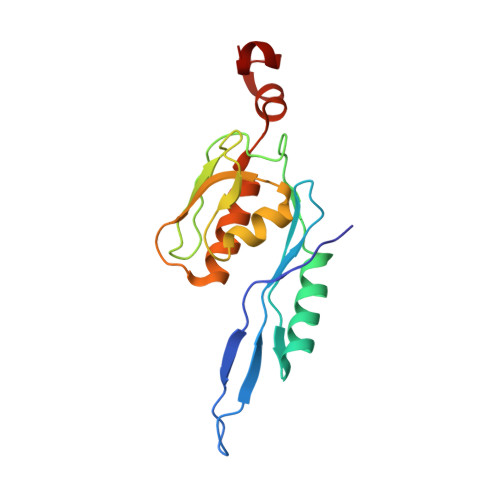
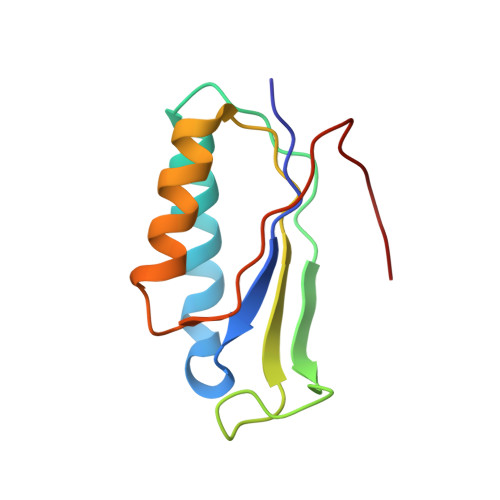
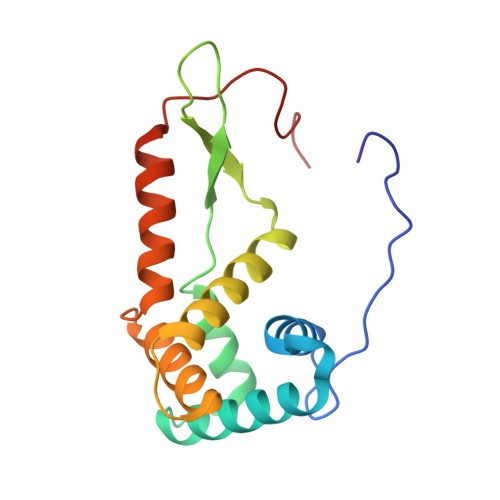
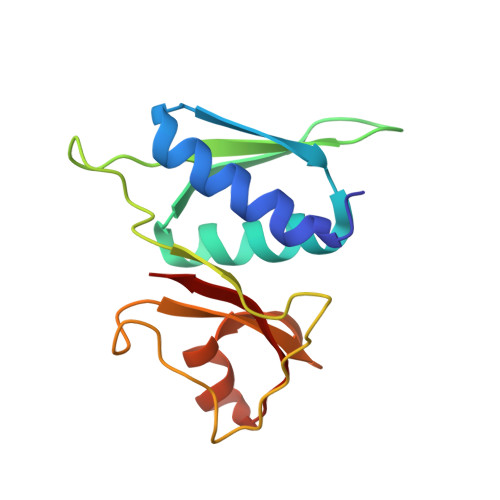
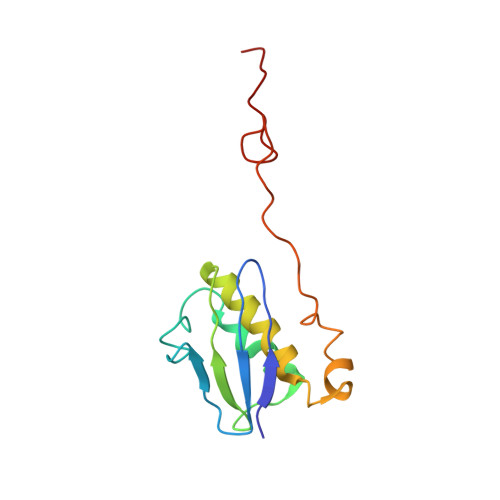
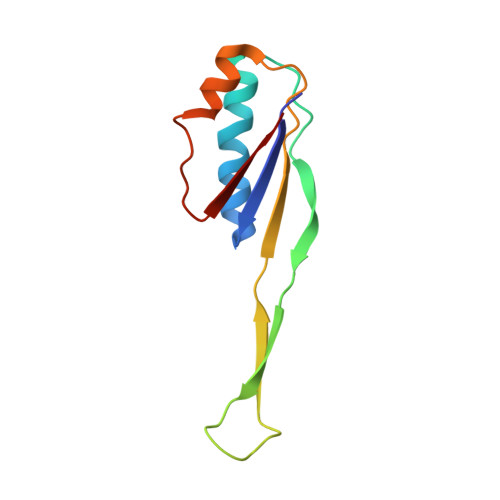
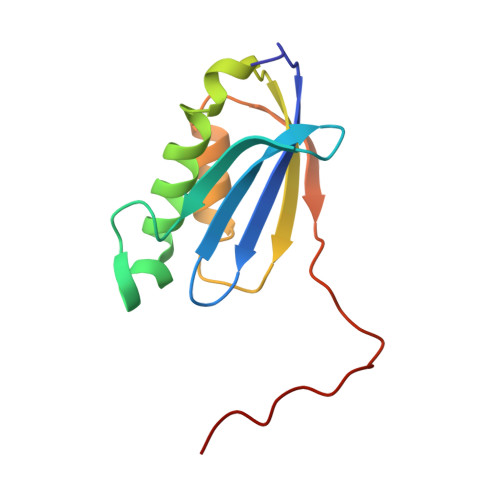
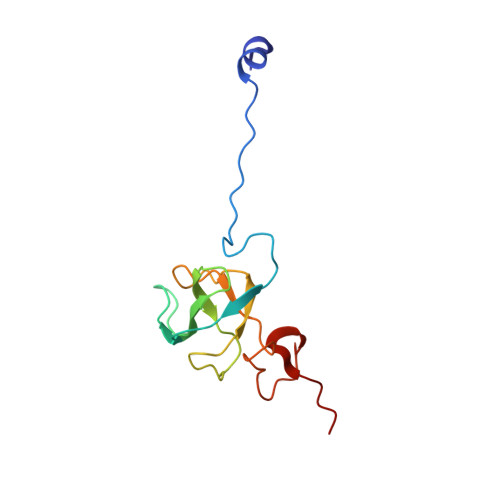
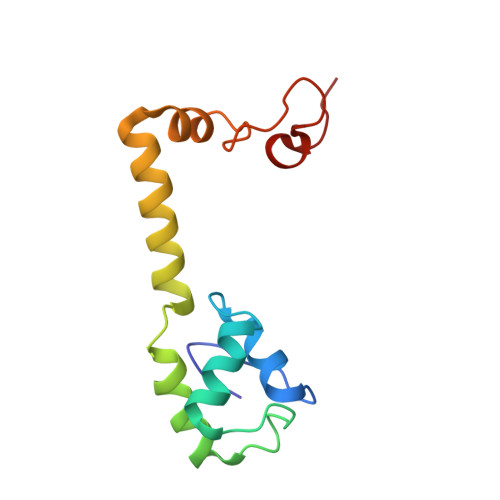
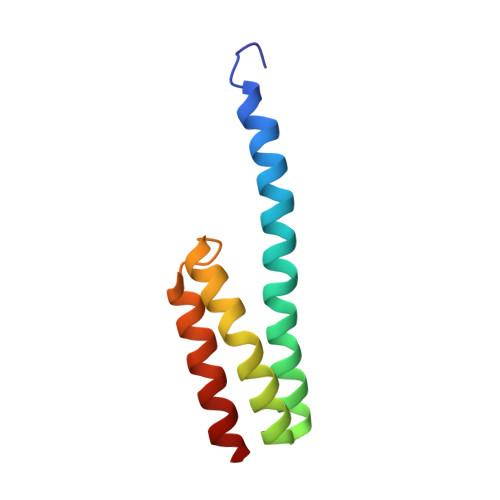
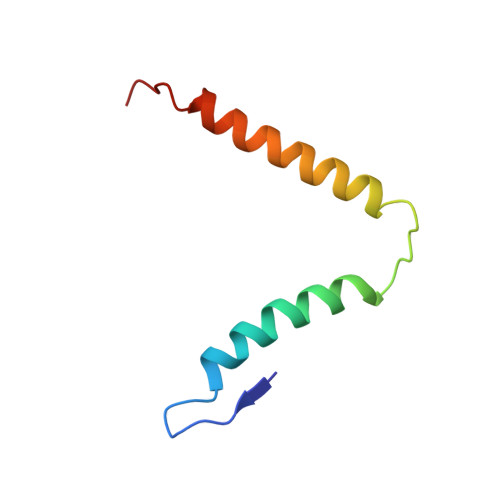
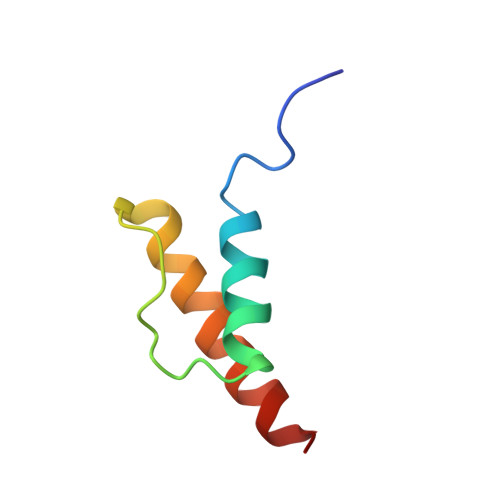
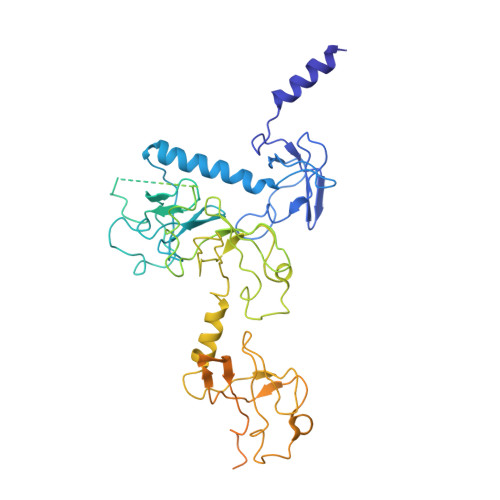
To survive under conditions of stress, such as nutrient deprivation, bacterial 70S ribosomes dimerize to form hibernating 100S particles 1 . In γ-proteobacteria, such as Escherichia coli, 100S formation requires the ribosome modulation factor (RMF) and the hibernation promoting factor (HPF) 2-4 . Here we present single-particle cryo-electron microscopy structures of hibernating 70S and 100S particles isolated from stationary-phase E. coli cells at 3.0 Å and 7.9 Å resolution, respectively. The structures reveal the binding sites for HPF and RMF as well as the unexpected presence of deacylated E-site transfer RNA and ribosomal protein bS1. HPF interacts with the anticodon-stem-loop of the E-tRNA and occludes the binding site for the messenger RNA as well as A- and P-site tRNAs. RMF facilitates stabilization of a compact conformation of bS1, which together sequester the anti-Shine-Dalgarno sequence of the 16S ribosomal RNA (rRNA), thereby inhibiting translation initiation. At the dimerization interface, the C-terminus of uS2 probes the mRNA entrance channel of the symmetry-related particle, thus suggesting that dimerization inactivates ribosomes by blocking the binding of mRNA within the channel. The back-to-back E. coli 100S arrangement is distinct from 100S particles observed previously in Gram-positive bacteria 5-8 , and reveals a unique role for bS1 in translation regulation.
- Institute for Biochemistry and Molecular Biology, University of Hamburg, Hamburg, Germany.
Organizational Affiliation: