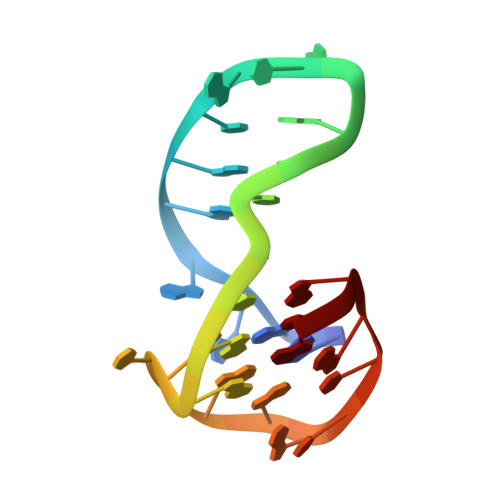
Major G-Quadruplex Form of HIV-1 LTR Reveals a (3 + 1) Folding Topology Containing a Stem-Loop.
Butovskaya, E., Heddi, B., Bakalar, B., Richter, S.N., Phan, A.T.(2018) J Am Chem Soc 140: 13654-13662
- PubMed: 30299955
- DOI: https://doi.org/10.1021/jacs.8b05332
- Primary Citation of Related Structures:
6H1K - PubMed Abstract:
Nucleic acids can form noncanonical four-stranded structures called G-quadruplexes. G-quadruplex-forming sequences are found in several genomes including human and viruses. Previous studies showed that the G-rich sequence located in the U3 promoter region of the HIV-1 long terminal repeat (LTR) folds into a set of dynamically interchangeable G-quadruplex structures. G-quadruplexes formed in the LTR could act as silencer elements to regulate viral transcription. Stabilization of LTR G-quadruplexes by G-quadruplex-specific ligands resulted in decreased viral production, suggesting the possibility of targeting viral G-quadruplex structures for antiviral purposes. Among all the G-quadruplexes formed in the LTR sequence, LTR-III was shown to be the major G-quadruplex conformation in vitro. Here we report the NMR structure of LTR-III in K + solution, revealing the formation of a unique quadruplex-duplex hybrid consisting of a three-layer (3 + 1) G-quadruplex scaffold, a 12-nt diagonal loop containing a conserved duplex-stem, a 3-nt lateral loop, a 1-nt propeller loop, and a V-shaped loop. Our structure showed several distinct features including a quadruplex-duplex junction, representing an attractive motif for drug targeting. The structure solved in this study may be used as a promising target to selectively impair the viral cycle.
- School of Physical and Mathematical Sciences , Nanyang Technological University , Singapore 637371 , Singapore.
Organizational Affiliation: