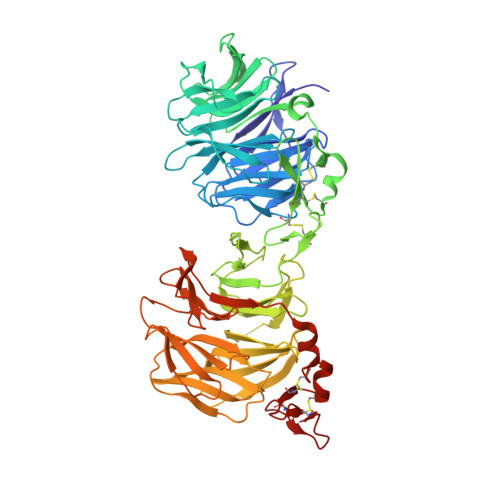
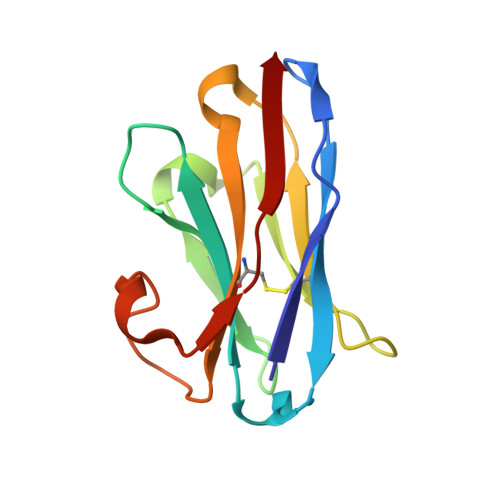
Anti-LRP5/6 VHHs promote differentiation of Wnt-hypersensitive intestinal stem cells.
Fenderico, N., van Scherpenzeel, R.C., Goldflam, M., Proverbio, D., Jordens, I., Kralj, T., Stryeck, S., Bass, T.Z., Hermans, G., Ullman, C., Aastrup, T., Gros, P., Maurice, M.M.(2019) Nat Commun 10: 365-365
- PubMed: 30664649
- DOI: https://doi.org/10.1038/s41467-018-08172-z
- Primary Citation of Related Structures:
6H15, 6H16 - PubMed Abstract:
Wnt-induced β-catenin-mediated transcription is a driving force for stem cell self-renewal during adult tissue homeostasis. Enhanced Wnt receptor expression due to mutational inactivation of the ubiquitin ligases RNF43/ZNRF3 recently emerged as a leading cause for cancer development. Consequently, targeting canonical Wnt receptors such as LRP5/6 holds great promise for treatment of such cancer subsets. Here, we employ CIS display technology to identify single-domain antibody fragments (VHH) that bind the LRP6 P3E3P4E4 region with nanomolar affinity and strongly inhibit Wnt3/3a-induced β-catenin-mediated transcription in cells, while leaving Wnt1 responses unaffected. Structural analysis reveal that individual VHHs variably employ divergent antigen-binding regions to bind a similar surface in the third β-propeller of LRP5/6, sterically interfering with Wnt3/3a binding. Importantly, anti-LRP5/6 VHHs block the growth of Wnt-hypersensitive Rnf43/Znrf3-mutant intestinal organoids through stem cell exhaustion and collective terminal differentiation. Thus, VHH-mediated targeting of LRP5/6 provides a promising differentiation-inducing strategy for treatment of Wnt-hypersensitive tumors.
- Oncode Institute and Department of Cell Biology, Center for Molecular Medicine, University Medical Center Utrecht, 3584 CX, Utrecht, The Netherlands.
Organizational Affiliation: