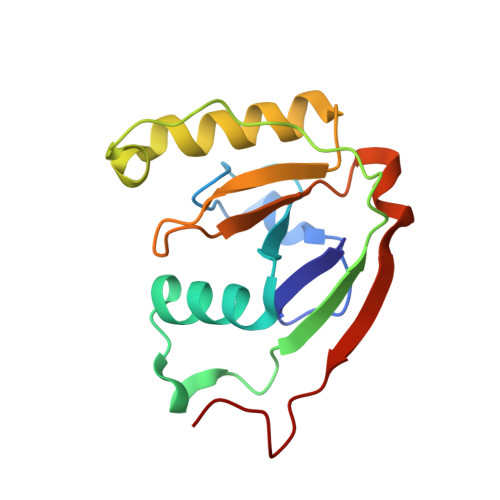
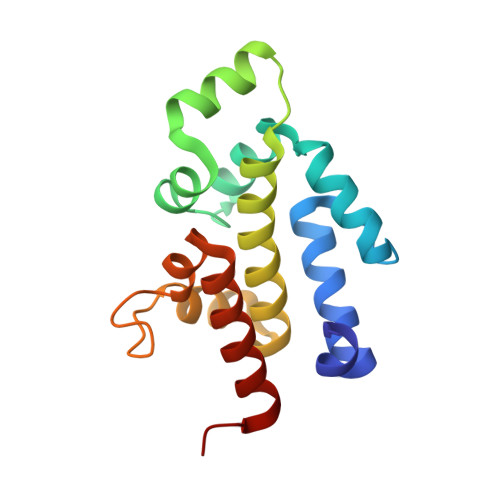
The RES domain toxins of RES-Xre toxin-antitoxin modules induce cell stasis by degrading NAD.
Skjerning, R.B., Senissar, M., Winther, K.S., Gerdes, K., Brodersen, D.E.(2019) Mol Microbiol 111: 221-236
- PubMed: 30315706
- DOI: https://doi.org/10.1111/mmi.14150
- Primary Citation of Related Structures:
6GW6 - PubMed Abstract:
Type II toxin-antitoxin (TA) modules, which are important cellular regulators in prokaryotes, usually encode two proteins, a toxin that inhibits cell growth and a nontoxic and labile inhibitor (antitoxin) that binds to and neutralizes the toxin. Here, we demonstrate that the res-xre locus from Photorhabdus luminescens and other bacterial species function as bona fide TA modules in Escherichia coli. The 2.2 Å crystal structure of the intact Pseudomonas putida RES-Xre TA complex reveals an unusual 2:4 stoichiometry in which a central RES toxin dimer binds two Xre antitoxin dimers. The antitoxin dimers each expose two helix-turn-helix DNA-binding domains of the Cro repressor type, suggesting the TA complex is capable of binding the upstream promoter sequence on DNA. The toxin core domain shows structural similarity to ADP-ribosylating enzymes such as diphtheria toxin but has an atypical NAD + -binding pocket suggesting an alternative function. We show that activation of the toxin in vivo causes a depletion of intracellular NAD + levels eventually leading to inhibition of cell growth in E. coli and inhibition of global macromolecular biosynthesis. Both structure and activity are unprecedented among bacterial TA systems, suggesting the functional scope of bacterial TA toxins is much wider than previously appreciated.
- Department of Biology, Centre for Bacterial Stress Response and Persistence (BASP), University of Copenhagen, Copenhagen, Denmark.
Organizational Affiliation: