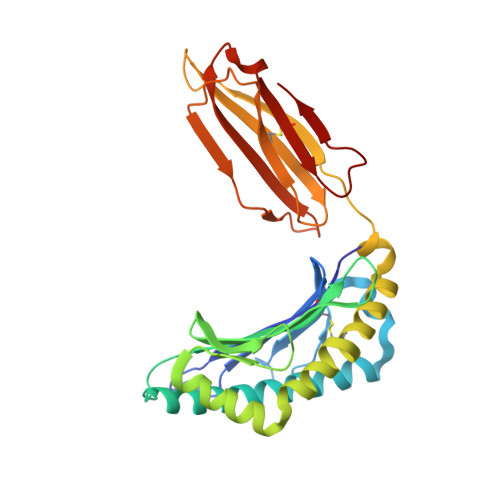
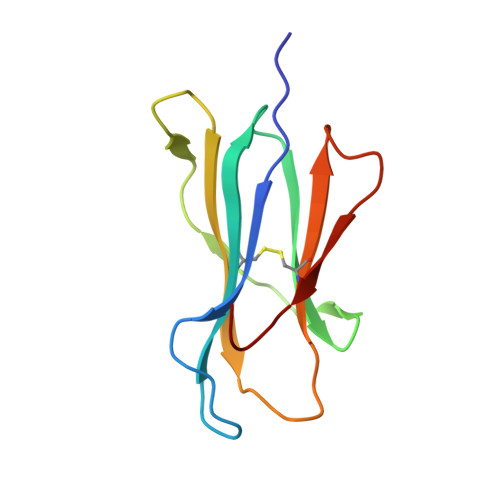
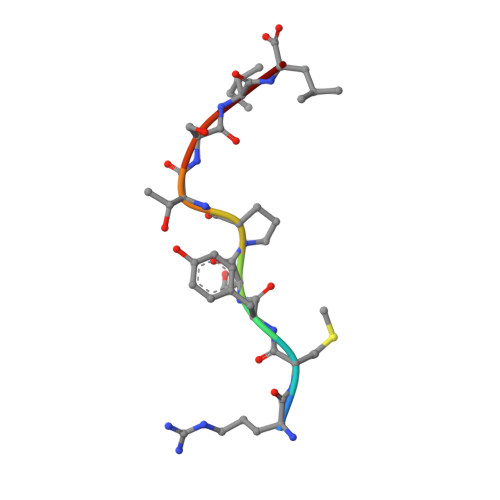
Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding.
Walters, L.C., Harlos, K., Brackenridge, S., Rozbesky, D., Barrett, J.R., Jain, V., Walter, T.S., O'Callaghan, C.A., Borrow, P., Toebes, M., Hansen, S.G., Sacha, J., Abdulhaqq, S., Greene, J.M., Fruh, K., Marshall, E., Picker, L.J., Jones, E.Y., McMichael, A.J., Gillespie, G.M.(2018) Nat Commun 9: 3137-3137
- PubMed: 30087334
- DOI: https://doi.org/10.1038/s41467-018-05459-z
- Primary Citation of Related Structures:
6GGM, 6GH1, 6GH4, 6GHN, 6GL1 - PubMed Abstract:
Through major histocompatibility complex class Ia leader sequence-derived (VL9) peptide binding and CD94/NKG2 receptor engagement, human leucocyte antigen E (HLA-E) reports cellular health to NK cells. Previous studies demonstrated a strong bias for VL9 binding by HLA-E, a preference subsequently supported by structural analyses. However, Mycobacteria tuberculosis (Mtb) infection and Rhesus cytomegalovirus-vectored SIV vaccinations revealed contexts where HLA-E and the rhesus homologue, Mamu-E, presented diverse pathogen-derived peptides to CD8 + T cells, respectively. Here we present crystal structures of HLA-E in complex with HIV and Mtb-derived peptides. We show that despite the presence of preferred primary anchor residues, HLA-E-bound peptides can adopt alternative conformations within the peptide binding groove. Furthermore, combined structural and mutagenesis analyses illustrate a greater tolerance for hydrophobic and polar residues in the primary pockets than previously appreciated. Finally, biochemical studies reveal HLA-E peptide binding and exchange characteristics with potential relevance to its alternative antigen presenting function in vivo.
- Nuffield Department of Medicine Research Building, Roosevelt Drive, Nuffield Department of Medicine, University of Oxford, Oxford, OX3 7FZ, UK.
Organizational Affiliation: