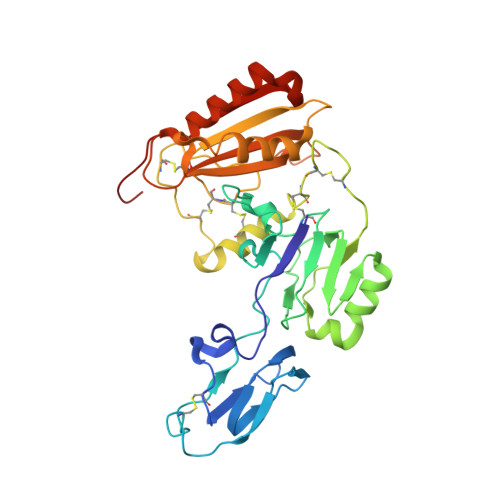
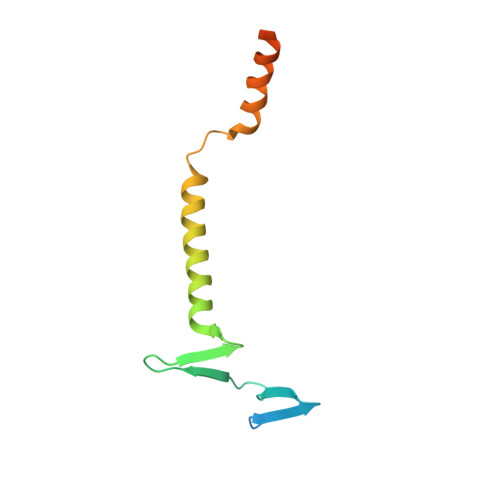
Structural assembly of the megadalton-sized receptor for intestinal vitamin B12uptake and kidney protein reabsorption.
Larsen, C., Etzerodt, A., Madsen, M., Skjodt, K., Moestrup, S.K., Andersen, C.B.F.(2018) Nat Commun 9: 5204-5204
- PubMed: 30523278
- DOI: https://doi.org/10.1038/s41467-018-07468-4
- Primary Citation of Related Structures:
6GJE - PubMed Abstract:
The endocytic receptor cubam formed by the 460-kDa protein cubilin and the 45-kDa transmembrane protein amnionless (AMN), is essential for intestinal vitamin B 12 (B 12 ) uptake and for protein (e.g. albumin) reabsorption from the kidney filtrate. Loss of function of any of the two components ultimately leads to serious B 12 deficiency and urinary protein loss in humans (Imerslund-Gräsbeck's syndrome, IGS). Here, we present the crystal structure of AMN in complex with the amino-terminal region of cubilin, revealing a sophisticated assembly of three cubilin subunits combining into a single intertwined β-helix domain that docks to a corresponding three-faced β-helix domain in AMN. This β-helix-β-helix association thereby anchors three ligand-binding cubilin subunits to the transmembrane AMN. Electron microscopy of full-length cubam reveals a 700-800 Å long tree-like structure with the potential of dimerization into an even larger complex. Furthermore, effects of known human mutations causing IGS are explained by the structural information.
- Department of Biomedicine, Aarhus University, 8000, Aarhus C, Denmark.
Organizational Affiliation: