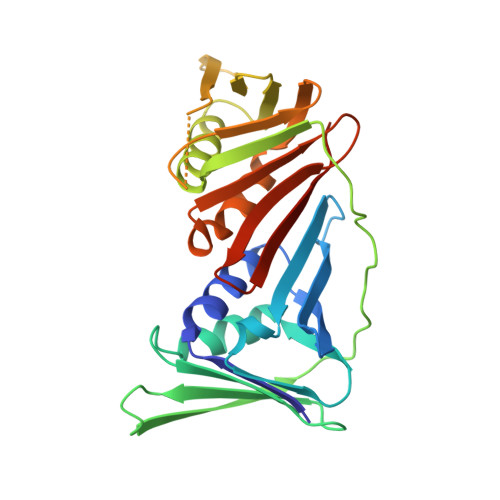
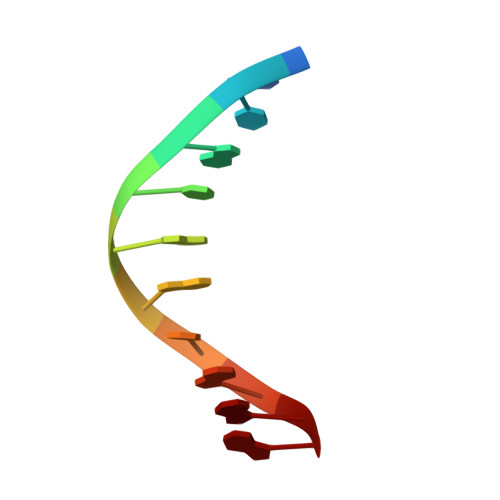
Structural basis of human PCNA sliding on DNA.
De March, M., Merino, N., Barrera-Vilarmau, S., Crehuet, R., Onesti, S., Blanco, F.J., De Biasio, A.(2017) Nat Commun 8: 13935-13935
- PubMed: 28071730
- DOI: https://doi.org/10.1038/ncomms13935
- Primary Citation of Related Structures:
6GIS - PubMed Abstract:
Sliding clamps encircle DNA and tether polymerases and other factors to the genomic template. However, the molecular mechanism of clamp sliding on DNA is unknown. Using crystallography, NMR and molecular dynamics simulations, here we show that the human clamp PCNA recognizes DNA through a double patch of basic residues within the ring channel, arranged in a right-hand spiral that matches the pitch of B-DNA. We propose that PCNA slides by tracking the DNA backbone via a 'cogwheel' mechanism based on short-lived polar interactions, which keep the orientation of the clamp invariant relative to DNA. Mutation of residues at the PCNA-DNA interface has been shown to impair the initiation of DNA synthesis by polymerase δ (pol δ). Therefore, our findings suggest that a clamp correctly oriented on DNA is necessary for the assembly of a replication-competent PCNA-pol δ holoenzyme.
- Structural Biology Laboratory, Elettra-Sincrotrone Trieste S.C.p.A., 34149 Trieste, Italy.
Organizational Affiliation: