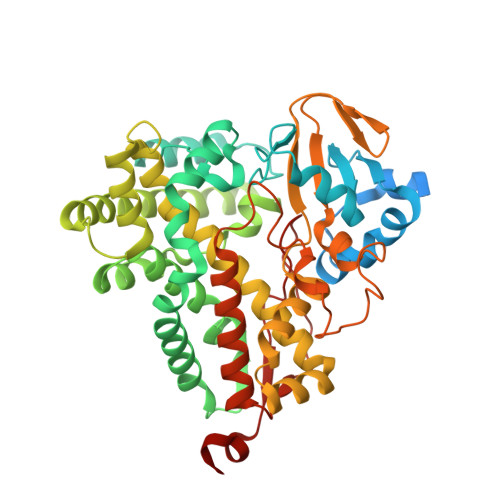
The crystal structure of P450-TT heme-domain provides the first structural insights into the versatile class VII P450s.
Tavanti, M., Porter, J.L., Levy, C.W., Gomez Castellanos, J.R., Flitsch, S.L., Turner, N.J.(2018) Biochem Biophys Res Commun 501: 846-850
- PubMed: 29738765
- DOI: https://doi.org/10.1016/j.bbrc.2018.05.014
- Primary Citation of Related Structures:
6GII - PubMed Abstract:
The first crystal structure of a class VII P450, CYP116B46 from Tepidiphilus thermophilus, has been solved at 1.9 Å resolution. The structure reveals overall conservation of the P450-fold and a water conduit around the I-helix. Active site residues have been identified and sequence comparisons have been made with other class VII enzymes. A structure similarity search demonstrated that the P450-TT structure is similar to enzymes capable of oxy-functionalization of fatty acids, terpenes, macrolides, steroids and statins. The insight gained from solving this structure will provide a guideline for future engineering and modelling studies on this catalytically promiscuous class of enzymes.
- Manchester Institute of Biotechnology (MIB), School of Chemistry, The University of Manchester, 131Princess Street, M1 7DN, Manchester, United Kingdom.
Organizational Affiliation: