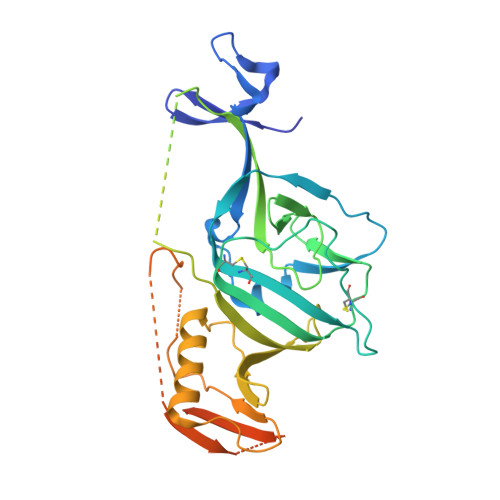
Structures of Ebola Virus Glycoprotein Complexes with Tricyclic Antidepressant and Antipsychotic Drugs.
Zhao, Y., Ren, J., Fry, E.E., Xiao, J., Townsend, A.R., Stuart, D.I.(2018) J Med Chem 61: 4938-4945
- PubMed: 29741894
- DOI: https://doi.org/10.1021/acs.jmedchem.8b00350
- Primary Citation of Related Structures:
6G95, 6G9B, 6G9I - PubMed Abstract:
A large number of Food and Drug Administration (FDA)-approved drugs have been found to inhibit the cell entry of Ebola virus (EBOV). However, since these drugs have various primary pharmacological targets, their mechanisms of action against EBOV remain largely unknown. We have previously shown that six FDA-approved drugs inhibit EBOV infection by interacting with and destabilizing the viral glycoprotein (GP). Here we show that antidepressants imipramine and clomipramine and antipsychotic drug thioridazine also directly interact with EBOV GP and determine the mode of interaction by crystallographic analysis of the complexes. The compounds bind within the same pocket as observed for other, chemically divergent complexes but with different binding modes. These details should be of value for the development of potent EBOV inhibitors.
- Division of Structural Biology , University of Oxford , The Henry Wellcome Building for Genomic Medicine , Headington, Oxford OX3 7BN , U.K.
Organizational Affiliation: