A GenoChemetic strategy for derivatization of the violacein natural product scaffold
Lai, H.E., Obled, A.M.C., Chee, S.M., Morgan, R.M., Sharma, S.V., Moore, S.J., Polizzi, K.M., Goss, R.J.M., Freemont, P.S.(2019) bioRxiv
Experimental Data Snapshot
Starting Model: experimental
View more details
(2019) bioRxiv
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Flavin-dependent L-tryptophan oxidase VioA | A [auth B], B [auth A] | 417 | Chromobacterium violaceum ATCC 12472 | Mutation(s): 0 Gene Names: vioA, CV_3274 EC: 1.4.3.23 | 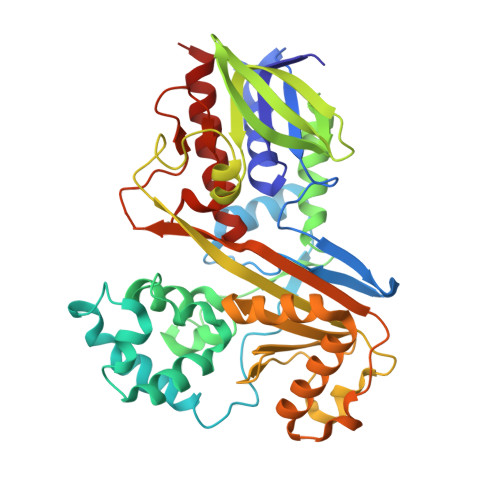 |
UniProt | |||||
Find proteins for Q9S3V1 (Chromobacterium violaceum (strain ATCC 12472 / DSM 30191 / JCM 1249 / CCUG 213 / NBRC 12614 / NCIMB 9131 / NCTC 9757 / MK)) Explore Q9S3V1 Go to UniProtKB: Q9S3V1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9S3V1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FAD Query on FAD | C [auth B], F [auth A] | FLAVIN-ADENINE DINUCLEOTIDE C27 H33 N9 O15 P2 VWWQXMAJTJZDQX-UYBVJOGSSA-N |  | ||
| FT6 Query on FT6 | D [auth B], G [auth A] | 6-FLUORO-L-TRYPTOPHAN C11 H11 F N2 O2 YMEXGEAJNZRQEH-VIFPVBQESA-N |  | ||
| MG Query on MG | E [auth B], H [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 151.478 | α = 90 |
| b = 174.322 | β = 90 |
| c = 93.821 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| xia2 | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Imperial College London | United Kingdom | President's PhD Scholarship |