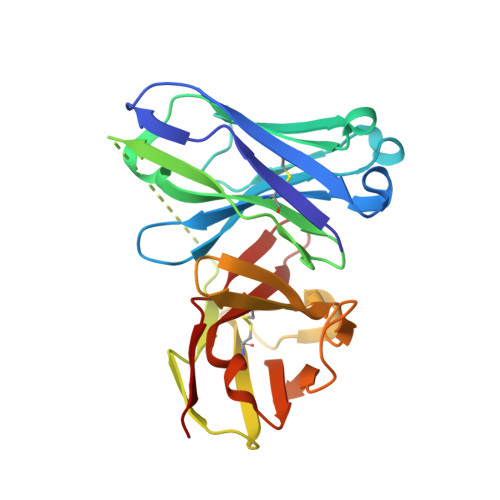
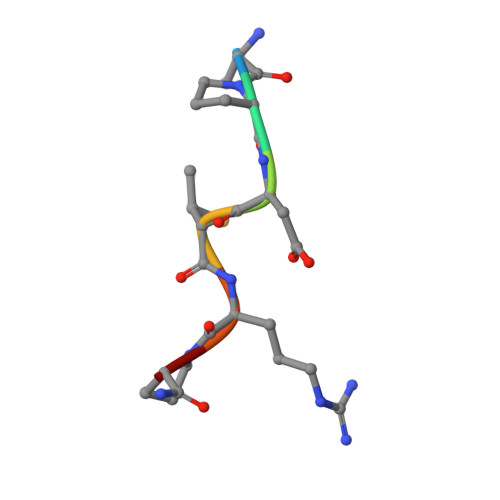
Structure-Based Design of Potent Tumor-Associated Antigens: Modulation of Peptide Presentation by Single-Atom O/S or O/Se Substitutions at the Glycosidic Linkage.
Companon, I., Guerreiro, A., Mangini, V., Castro-Lopez, J., Escudero-Casao, M., Avenoza, A., Busto, J.H., Castillon, S., Jimenez-Barbero, J., Asensio, J.L., Jimenez-Oses, G., Boutureira, O., Peregrina, J.M., Hurtado-Guerrero, R., Fiammengo, R., Bernardes, G.J.L., Corzana, F.(2019) J Am Chem Soc 141: 4063-4072
- PubMed: 30726084
- DOI: https://doi.org/10.1021/jacs.8b13503
- Primary Citation of Related Structures:
5N7B, 6FRJ - PubMed Abstract:
GalNAc-glycopeptides derived from mucin MUC1 are an important class of tumor-associated antigens. α- O-glycosylation forces the peptide to adopt an extended conformation in solution, which is far from the structure observed in complexes with a model anti-MUC1 antibody. Herein, we propose a new strategy for designing potent antigen mimics based on modulating peptide/carbohydrate interactions by means of O → S/Se replacement at the glycosidic linkage. These minimal chemical modifications bring about two key structural changes to the glycopeptide. They increase the carbohydrate-peptide distance and change the orientation and dynamics of the glycosidic linkage. As a result, the peptide acquires a preorganized and optimal structure suited for antibody binding. Accordingly, these new glycopeptides display improved binding toward a representative anti-MUC1 antibody relative to the native antigens. To prove the potential of these glycopeptides as tumor-associated MUC1 antigen mimics, the derivative bearing the S-glycosidic linkage was conjugated to gold nanoparticles and tested as an immunogenic formulation in mice without any adjuvant, which resulted in a significant humoral immune response. Importantly, the mice antisera recognize cancer cells in biopsies of breast cancer patients with high selectivity. This finding demonstrates that the antibodies elicited against the mimetic antigen indeed recognize the naturally occurring antigen in its physiological context. Clinically, the exploitation of tumor-associated antigen mimics may contribute to the development of cancer vaccines and to the improvement of cancer diagnosis based on anti-MUC1 antibodies. The methodology presented here is of general interest for applications because it may be extended to modulate the affinity of biologically relevant glycopeptides toward their receptors.
- Departamento de Química , Universidad de La Rioja , Centro de Investigación en Síntesis Química , 26006 Logroño , Spain.
Organizational Affiliation: