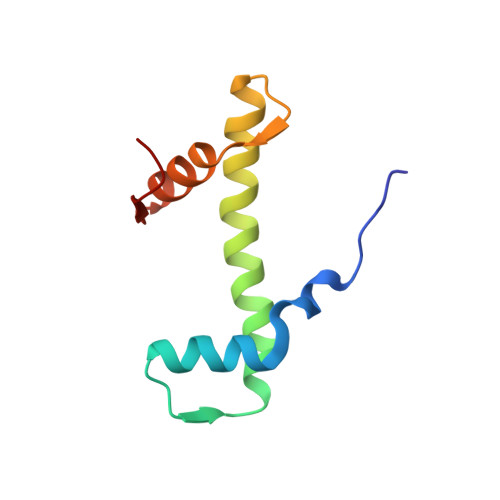
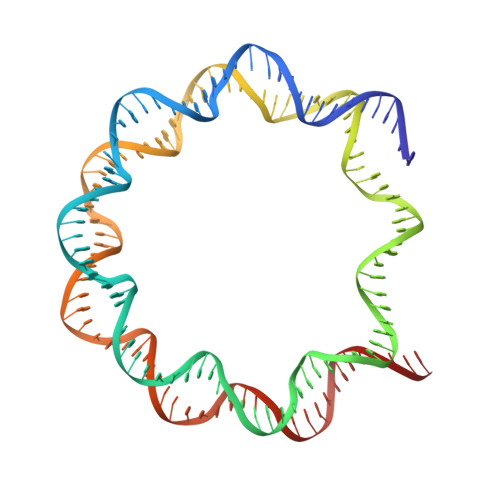
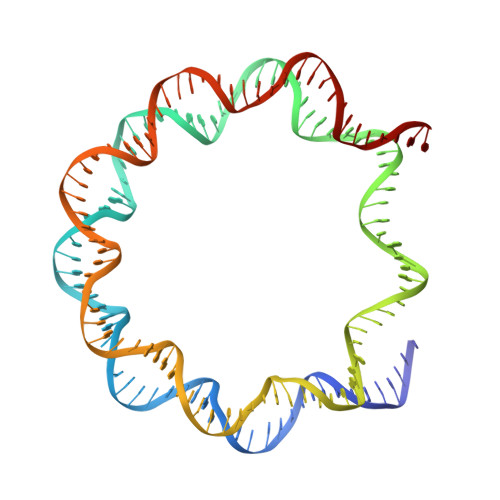
Structural rearrangements of the histone octamer translocate DNA.
Bilokapic, S., Strauss, M., Halic, M.(2018) Nat Commun 9: 1330-1330
- PubMed: 29626188
- DOI: https://doi.org/10.1038/s41467-018-03677-z
- Primary Citation of Related Structures:
6FQ5, 6FQ6, 6FQ8 - PubMed Abstract:
Nucleosomes, the basic unit of chromatin, package and regulate expression of eukaryotic genomes. Nucleosomes are highly dynamic and are remodeled with the help of ATP-dependent remodeling factors. Yet, the mechanism of DNA translocation around the histone octamer is poorly understood. In this study, we present several nucleosome structures showing histone proteins and DNA in different organizational states. We observe that the histone octamer undergoes conformational changes that distort the overall nucleosome structure. As such, rearrangements in the histone core α-helices and DNA induce strain that distorts and moves DNA at SHL 2. Distortion of the nucleosome structure detaches histone α-helices from the DNA, leading to their rearrangement and DNA translocation. Biochemical assays show that cross-linked histone octamers are immobilized on DNA, indicating that structural changes in the octamer move DNA. This intrinsic plasticity of the nucleosome is exploited by chromatin remodelers and might be used by other chromatin machineries.
- Gene Ceter, Department of Biochemistry, University of Munich LMU, 81377, Munich, Germany.
Organizational Affiliation: