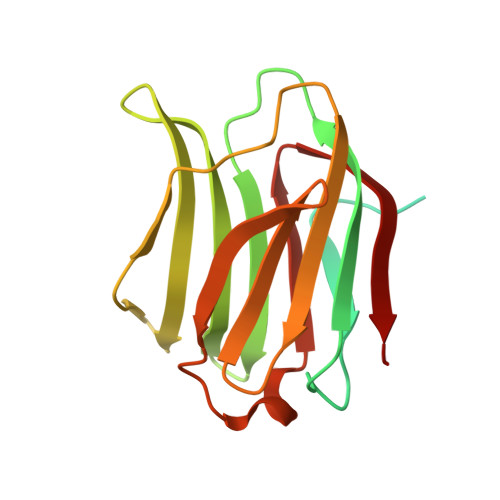
Crystallization of a human galectin-3 variant with two ordered segments in the shortened N-terminal tail.
Flores-Ibarra, A., Vertesy, S., Medrano, F.J., Gabius, H.J., Romero, A.(2018) Sci Rep 8: 9835-9835
- PubMed: 29959397
- DOI: https://doi.org/10.1038/s41598-018-28235-x
- Primary Citation of Related Structures:
6FOF - PubMed Abstract:
Among members of the family of adhesion/growth-regulatory galectins, galectin-3 (Gal-3) bears a unique modular architecture. A N-terminal tail (NT) consisting of the N-terminal segment (NTS) and nine collagen-like repeats is linked to the canonical lectin domain. In contrast to bivalent proto- and tandem-repeat-type galectins, Gal-3 is monomeric in solution, capable to self-associate in the presence of bi- to multivalent ligands, and the NTS is involved in cellular compartmentalization. Since no crystallographic information on Gal-3 beyond the lectin domain is available, we used a shortened variant with NTS and repeats VII-IX. This protein crystallized as tetramers with contacts between the lectin domains. The region from Tyr101 (in repeat IX) to Leu114 (in the CRD) formed a hairpin. The NTS extends the canonical β-sheet of F1-F5 strands with two new β-strands on the F face. Together, crystallographic and SAXS data reveal a mode of intramolecular structure building involving the highly flexible Gal-3's NT.
- Department of Structural and Chemical Biology, Centro de Investigaciones Biológicas, CSIC, Ramiro de Maeztu 9, 28040, Madrid, Spain.
Organizational Affiliation: