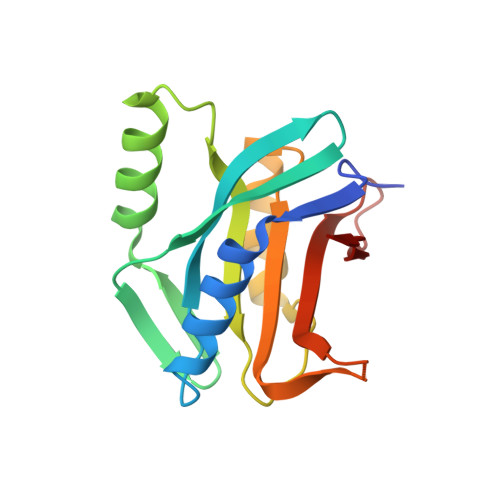
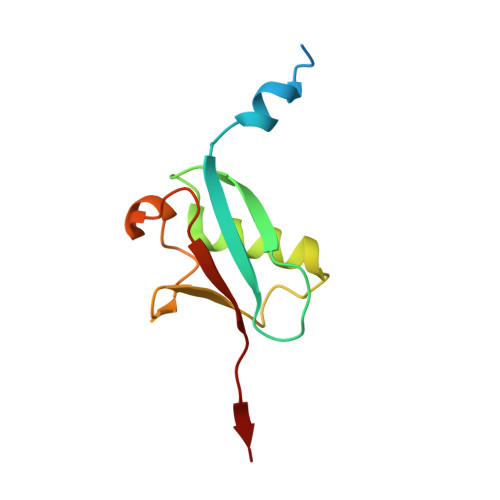
Rpn11-mediated ubiquitin processing in an ancestral archaeal ubiquitination system.
Fuchs, A.C.D., Maldoner, L., Wojtynek, M., Hartmann, M.D., Martin, J.(2018) Nat Commun 9: 2696-2696
- PubMed: 30002364
- DOI: https://doi.org/10.1038/s41467-018-05198-1
- Primary Citation of Related Structures:
6FJ7, 6FJU, 6FJV, 6FNN, 6FNO - PubMed Abstract:
While protein ubiquitination was long believed to be a truly eukaryotic feature, recently sequenced genomes revealed complete ubiquitin (Ub) modification operons in archaea. Here, we present the structural and mechanistic characterization of an archaeal Rpn11 deubiquitinase from Caldiarchaeum subterraneum, CsRpn11, and its role in the processing of CsUb precursor and ubiquitinated proteins. CsRpn11 activity is affected by the catalytic metal ion type, small molecule inhibitors, sequence characteristics at the cleavage site, and the folding state of CsUb-conjugated proteins. Comparison of CsRpn11 and CsRpn11-CsUb crystal structures reveals a crucial conformational switch in the CsRpn11 Ins-1 site, which positions CsUb for catalysis. The presence of this transition in a primordial soluble Rpn11 thus predates the evolution of eukaryotic Rpn11 immobilized in the proteasomal lid. Complementing phylogenetic studies, which designate CsRpn11 and CsUb as close homologs of the respective eukaryotic proteins, our results provide experimental support for an archaeal origin of protein ubiquitination.
- Department of Protein Evolution, Max Planck Institute for Developmental Biology, Max-Planck-Ring 5, 72076, Tübingen, Germany.
Organizational Affiliation: