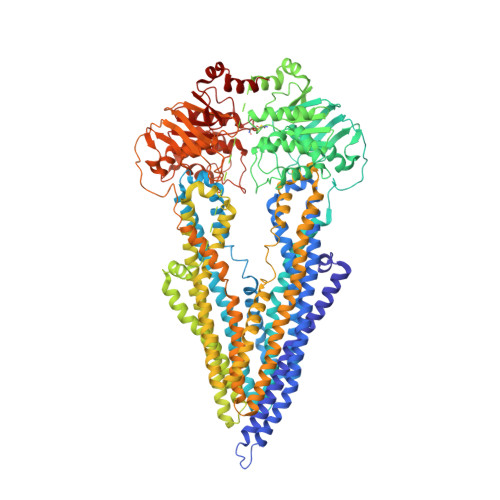
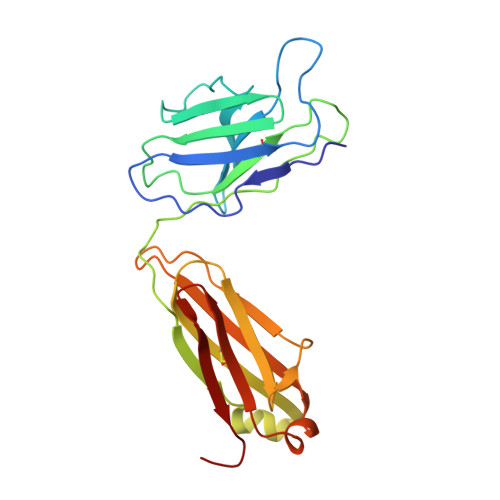
Structure of a zosuquidar and UIC2-bound human-mouse chimeric ABCB1.
Alam, A., Kung, R., Kowal, J., McLeod, R.A., Tremp, N., Broude, E.V., Roninson, I.B., Stahlberg, H., Locher, K.P.(2018) Proc Natl Acad Sci U S A 115: E1973-E1982
- PubMed: 29440498
- DOI: https://doi.org/10.1073/pnas.1717044115
- Primary Citation of Related Structures:
6FN1, 6FN4 - PubMed Abstract:
The multidrug transporter ABCB1 (P-glycoprotein) is an ATP-binding cassette transporter that has a key role in protecting tissues from toxic insult and contributes to multidrug extrusion from cancer cells. Here, we report the near-atomic resolution cryo-EM structure of nucleotide-free ABCB1 trapped by an engineered disulfide cross-link between the nucleotide-binding domains (NBDs) and bound to the antigen-binding fragment of the human-specific inhibitory antibody UIC2 and to the third-generation ABCB1 inhibitor zosuquidar. Our structure reveals the transporter in an occluded conformation with a central, enclosed, inhibitor-binding pocket lined by residues from all transmembrane (TM) helices of ABCB1. The pocket spans almost the entire width of the lipid membrane and is occupied exclusively by two closely interacting zosuquidar molecules. The external, conformational epitope facilitating UIC2 binding is also visualized, providing a basis for its inhibition of substrate efflux. Additional cryo-EM structures suggest concerted movement of TM helices from both halves of the transporters associated with closing the NBD gap, as well as zosuquidar binding. Our results define distinct recognition interfaces of ABCB1 inhibitory agents, which may be exploited for therapeutic purposes.
- Institute of Molecular Biology and Biophysics, ETH Zürich, 8093 Zürich, Switzerland.
Organizational Affiliation: