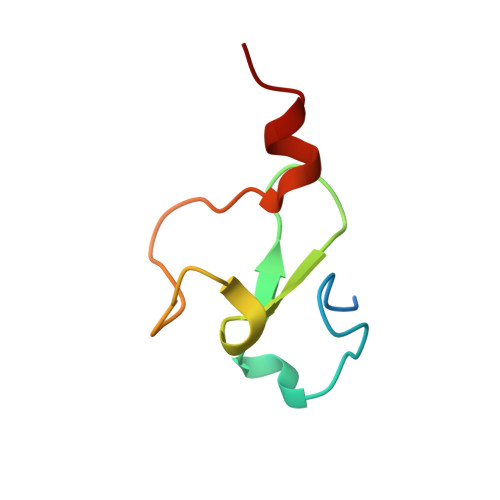
Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
Amato, A., Lucas, X., Bortoluzzi, A., Wright, D., Ciulli, A.(2018) ACS Chem Biol 13: 915-921
- PubMed: 29529862
- DOI: https://doi.org/10.1021/acschembio.7b01093
- Primary Citation of Related Structures:
6FAP, 6FHQ, 6FHU, 6FI0, 6FI1, 6FKP - PubMed Abstract:
Plant homeodomain (PHD) zinc fingers are histone reader domains that are often associated with human diseases. Despite this, they constitute a poorly targeted class of readers, suggesting low ligandability. Here, we describe a successful fragment-based campaign targeting PHD fingers from the proteins BAZ2A and BAZ2B as model systems. We validated a pool of in silico fragments both biophysically and structurally and solved the first crystal structures of PHD zinc fingers in complex with fragments bound to an anchoring pocket at the histone binding site. The best-validated hits were found to displace a histone H3 tail peptide in competition assays. This work identifies new chemical scaffolds that provide suitable starting points for future ligand optimization using structure-guided approaches. The demonstrated ligandability of the PHD reader domains could pave the way for the development of chemical probes to drug this family of epigenetic readers.
- Division of Biological Chemistry and Drug Discovery, School of Life Sciences , University of Dundee , James Black Centre, Dow Street , Dundee DD1 5EH , United Kingdom.
Organizational Affiliation: