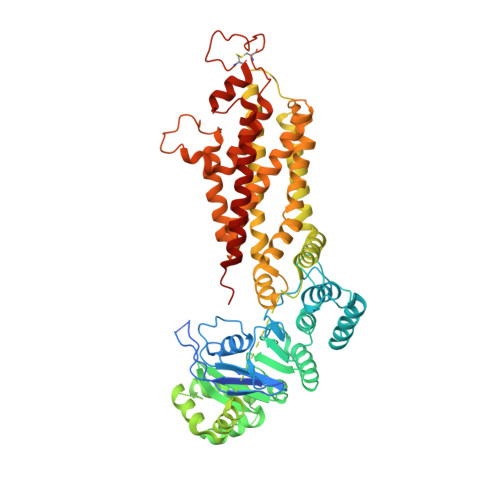
Structural basis of small-molecule inhibition of human multidrug transporter ABCG2.
Jackson, S.M., Manolaridis, I., Kowal, J., Zechner, M., Taylor, N.M.I., Bause, M., Bauer, S., Bartholomaeus, R., Bernhardt, G., Koenig, B., Buschauer, A., Stahlberg, H., Altmann, K.H., Locher, K.P.(2018) Nat Struct Mol Biol 25: 333-340
- PubMed: 29610494
- DOI: https://doi.org/10.1038/s41594-018-0049-1
- Primary Citation of Related Structures:
6ETI, 6FEQ, 6FFC, 6HIJ - PubMed Abstract:
ABCG2 is an ATP-binding cassette (ABC) transporter that protects tissues against xenobiotics, affects the pharmacokinetics of drugs and contributes to multidrug resistance. Although many inhibitors and modulators of ABCG2 have been developed, understanding their structure-activity relationship requires high-resolution structural insight. Here, we present cryo-EM structures of human ABCG2 bound to synthetic derivatives of the fumitremorgin C-related inhibitor Ko143 or the multidrug resistance modulator tariquidar. Both compounds are bound to the central, inward-facing cavity of ABCG2, blocking access for substrates and preventing conformational changes required for ATP hydrolysis. The high resolutions allowed for de novo building of the entire transporter and also revealed tightly bound phospholipids and cholesterol interacting with the lipid-exposed surface of the transmembrane domains (TMDs). Extensive chemical modifications of the Ko143 scaffold combined with in vitro functional analyses revealed the details of ABCG2 interactions with this compound family and provide a basis for the design of novel inhibitors and modulators.
- Institute of Molecular Biology and Biophysics, Department of Biology, ETH Zurich, Zurich, Switzerland.
Organizational Affiliation: