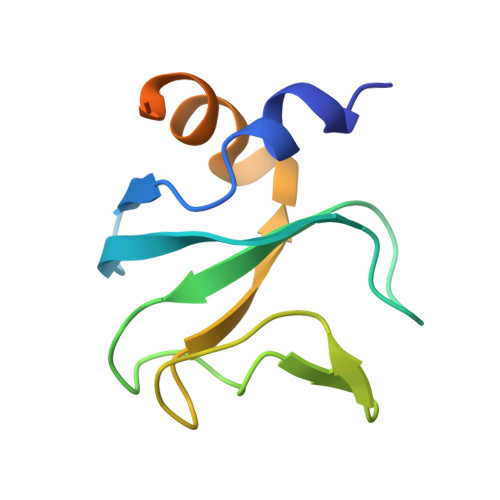
Structure-Function Relationship of the Bik1-Bim1 Complex.
Stangier, M.M., Kumar, A., Chen, X., Farcas, A.M., Barral, Y., Steinmetz, M.O.(2018) Structure 26: 607-618.e4
- PubMed: 29576319
- DOI: https://doi.org/10.1016/j.str.2018.03.003
- Primary Citation of Related Structures:
6FC5, 6FC6 - PubMed Abstract:
In budding yeast, the microtubule plus-end tracking proteins Bik1 (CLIP-170) and Bim1 (EB1) form a complex that interacts with partners involved in spindle positioning, including Stu2 and Kar9. Here, we show that the CAP-Gly and coiled-coil domains of Bik1 interact with the C-terminal ETF peptide of Bim1 and the C-terminal tail region of Stu2, respectively. The crystal structures of the CAP-Gly domain of Bik1 (Bik1CG) alone and in complex with an ETF peptide revealed unique, functionally relevant CAP-Gly elements, establishing Bik1CG as a specific C-terminal phenylalanine recognition domain. Unlike the mammalian CLIP-170-EB1 complex, Bik1-Bim1 forms ternary complexes with the EB1-binding motifs SxIP and LxxPTPh, which are present in diverse proteins, including Kar9. Perturbation of the Bik1-Bim1 interaction in vivo affected Bik1 localization and astral microtubule length. Our results provide insight into the role of the Bik1-Bim1 interaction for cell division, and demonstrate that the CLIP-170-EB1 module is evolutionarily flexible.
- Laboratory of Biomolecular Research, Division of Biology and Chemistry, Paul Scherrer Institut, 5232 Villigen PSI, Switzerland.
Organizational Affiliation: