Crystal structure of DARPin-DARPin rigid fusion, variant DD_D12_10_47 in complex JNK1a1 and JIP1 peptide
Wu, Y., Mittl, P.R., Honegger, A., Batyuk, A., Plueckthun, A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DD_D12_10_47 | 325 | synthetic construct | Mutation(s): 0 | 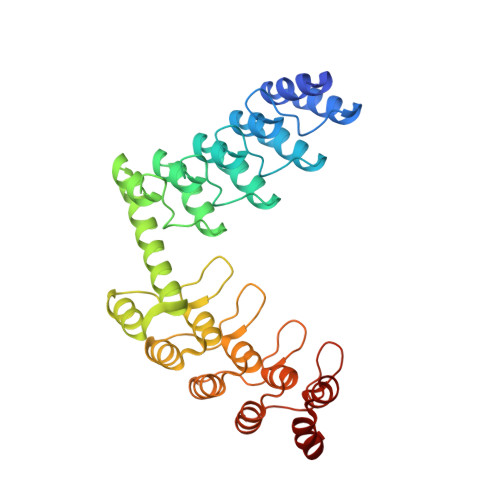 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Mitogen-activated protein kinase 8 | 373 | Homo sapiens | Mutation(s): 0 Gene Names: MAPK8, JNK1, PRKM8, SAPK1, SAPK1C EC: 2.7.11.24 | 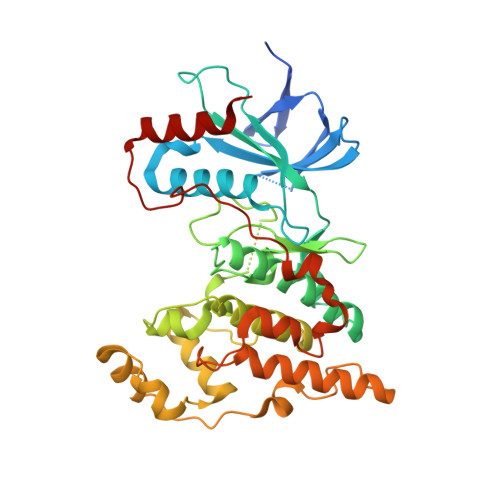 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P45983 (Homo sapiens) Explore P45983 Go to UniProtKB: P45983 | |||||
PHAROS: P45983 GTEx: ENSG00000107643 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P45983 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| C-Jun-amino-terminal kinase-interacting protein 1 | 11 | Mus musculus | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for Q9WVI9 (Mus musculus) Explore Q9WVI9 Go to UniProtKB: Q9WVI9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9WVI9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 76.995 | α = 90 |
| b = 76.995 | β = 90 |
| c = 330.725 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Swiss National Science Foundation | Switzerland | 310030B_166676 |
| European Research Council | NEXTBINDERS |