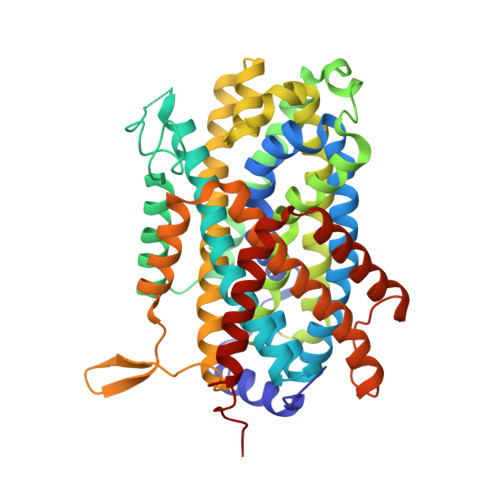
Structural basis for amino acid transport by the CAT family of SLC7 transporters.
Jungnickel, K.E.J., Parker, J.L., Newstead, S.(2018) Nat Commun 9: 550-550
- PubMed: 29416041
- DOI: https://doi.org/10.1038/s41467-018-03066-6
- Primary Citation of Related Structures:
5OQT, 6F34 - PubMed Abstract:
Amino acids play essential roles in cell biology as regulators of metabolic pathways. Arginine in particular is a major signalling molecule inside the cell, being a precursor for both l-ornithine and nitric oxide (NO) synthesis and a key regulator of the mTORC1 pathway. In mammals, cellular arginine availability is determined by members of the solute carrier (SLC) 7 family of cationic amino acid transporters. Whereas CAT-1 functions to supply cationic amino acids for cellular metabolism, CAT-2A and -2B are required for macrophage activation and play important roles in regulating inflammation. Here, we present the crystal structure of a close homologue of the mammalian CAT transporters that reveals how these proteins specifically recognise arginine. Our structural and functional data provide a model for cationic amino acid transport in mammalian cells and reveals mechanistic insights into proton-coupled, sodium-independent amino acid transport in the wider APC superfamily.
- Department of Biochemistry, University of Oxford, South Parks Road, Oxford, OX1 3QU, UK.
Organizational Affiliation: