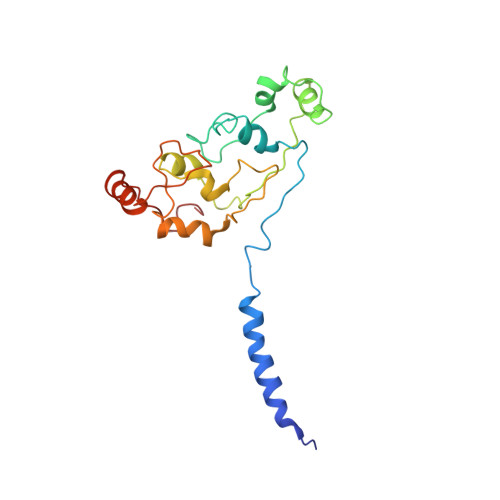
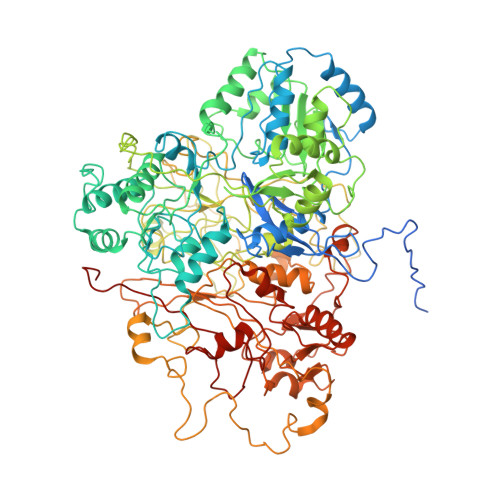
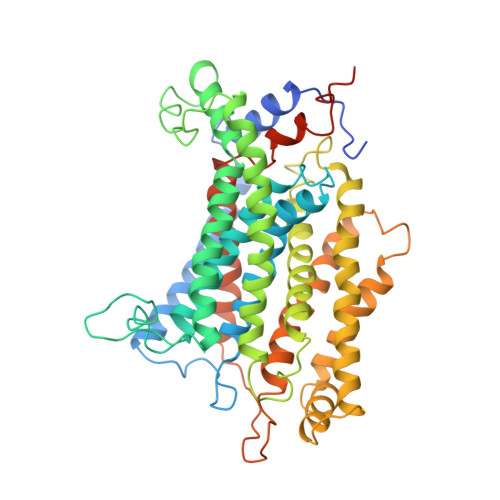
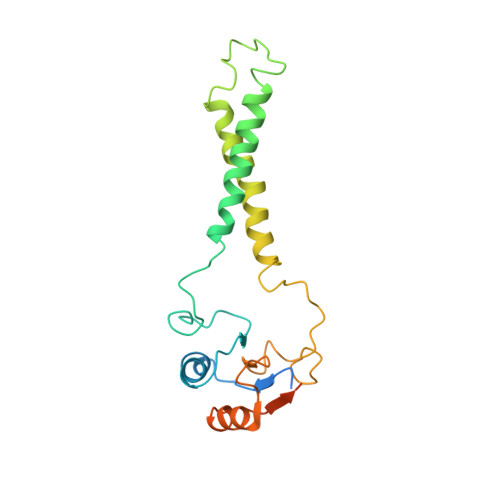
Structural basis for energy transduction by respiratory alternative complex III.
Sousa, J.S., Calisto, F., Langer, J.D., Mills, D.J., Refojo, P.N., Teixeira, M., Kuhlbrandt, W., Vonck, J., Pereira, M.M.(2018) Nat Commun 9: 1728-1728
- PubMed: 29712914
- DOI: https://doi.org/10.1038/s41467-018-04141-8
- Primary Citation of Related Structures:
6F0K - PubMed Abstract:
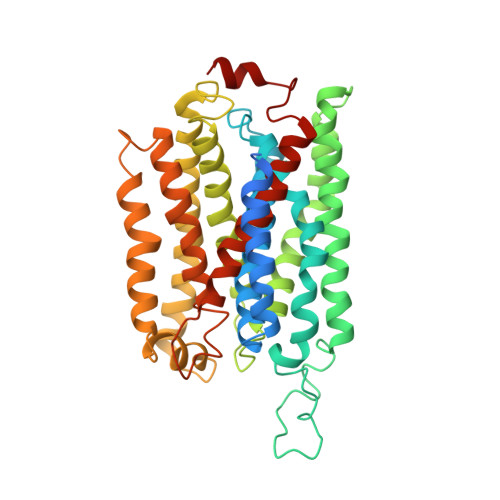
Electron transfer in respiratory chains generates the electrochemical potential that serves as energy source for the cell. Prokaryotes can use a wide range of electron donors and acceptors and may have alternative complexes performing the same catalytic reactions as the mitochondrial complexes. This is the case for the alternative complex III (ACIII), a quinol:cytochrome c/HiPIP oxidoreductase. In order to understand the catalytic mechanism of this respiratory enzyme, we determined the structure of ACIII from Rhodothermus marinus at 3.9 Å resolution by single-particle cryo-electron microscopy. ACIII presents a so-far unique structure, for which we establish the arrangement of the cofactors (four iron-sulfur clusters and six c-type hemes) and propose the location of the quinol-binding site and the presence of two putative proton pathways in the membrane. Altogether, this structure provides insights into a mechanism for energy transduction and introduces ACIII as a redox-driven proton pump.
- Department of Structural Biology, Max Planck Institute of Biophysics, Max-von-Laue Str. 3, 60438, Frankfurt am Main, Germany.
Organizational Affiliation: