Conceptional Design of Self-Assembling Bisubstrate-like Inhibitors of Protein Kinase A Resulting in a Boronic Acid Glutamate Linkage
Mueller, J.M., Kirschner, R., Geyer, A., Klebe, G.(2019) ACS Omega
Experimental Data Snapshot
(2019) ACS Omega
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| cAMP-dependent protein kinase catalytic subunit alpha | 353 | Cricetulus griseus | Mutation(s): 0 Gene Names: PRKACA EC: 2.7.11.11 | 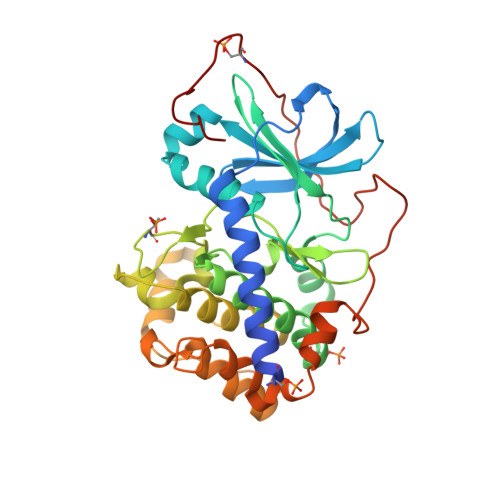 | |
UniProt | |||||
Find proteins for P25321 (Cricetulus griseus) Explore P25321 Go to UniProtKB: P25321 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P25321 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| cAMP-dependent protein kinase inhibitor | B [auth E] | 18 | Cricetulus griseus | Mutation(s): 0 | 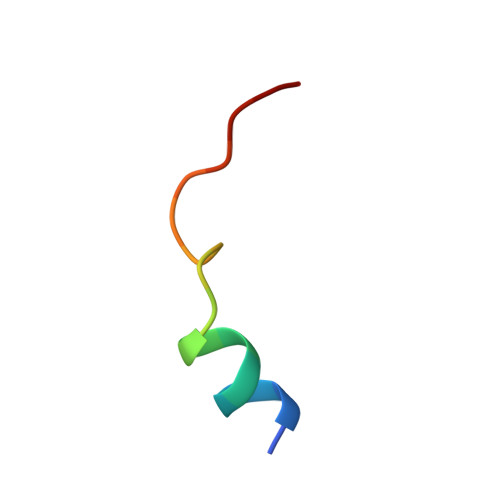 |
UniProt | |||||
Find proteins for G3HK48 (Cricetulus griseus) Explore G3HK48 Go to UniProtKB: G3HK48 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | G3HK48 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| AMP Query on AMP | C [auth A] | ADENOSINE MONOPHOSPHATE C10 H14 N5 O7 P UDMBCSSLTHHNCD-KQYNXXCUSA-N |  | ||
| RIP Query on RIP | F [auth E] | beta-D-ribopyranose C5 H10 O5 SRBFZHDQGSBBOR-TXICZTDVSA-N |  | ||
| MPD Query on MPD | D [auth A], E [auth A] | (4S)-2-METHYL-2,4-PENTANEDIOL C6 H14 O2 SVTBMSDMJJWYQN-YFKPBYRVSA-N |  | ||
| Modified Residues 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| SEP Query on SEP | A | L-PEPTIDE LINKING | C3 H8 N O6 P |  | SER |
| TPO Query on TPO | A | L-PEPTIDE LINKING | C4 H10 N O6 P |  | THR |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 64.939 | α = 90 |
| b = 80.327 | β = 90 |
| c = 85.002 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XDS | data scaling |
| PHASER | phasing |
| Coot | model building |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Loewe Corporation | Germany | -- |