Structural Basis of Splicing Modulation by Antitumor Macrolide Compounds.
Cretu, C., Agrawal, A.A., Cook, A., Will, C.L., Fekkes, P., Smith, P.G., Luhrmann, R., Larsen, N., Buonamici, S., Pena, V.(2018) Mol Cell 70: 265-273.e8
- PubMed: 29656923
- DOI: https://doi.org/10.1016/j.molcel.2018.03.011
- Primary Citation of Related Structures:
6EN4 - PubMed Abstract:
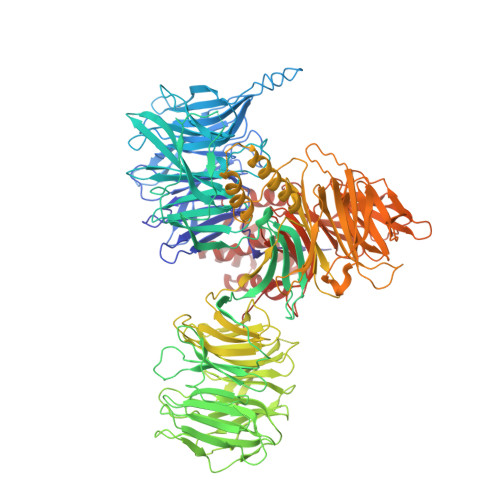
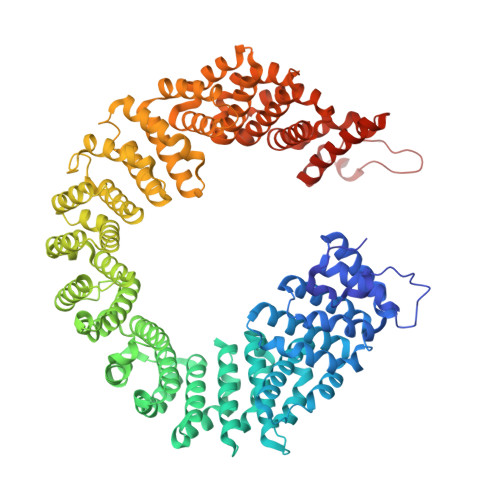
SF3B is a multi-protein complex essential for branch site (BS) recognition and selection during pre-mRNA splicing. Several splicing modulators with antitumor activity bind SF3B and thereby modulate splicing. Here we report the crystal structure of a human SF3B core in complex with pladienolide B (PB), a macrocyclic splicing modulator and potent inhibitor of tumor cell proliferation. PB stalls SF3B in an open conformation by acting like a wedge within a hinge, modulating SF3B's transition to the closed conformation needed to form the BS adenosine-binding pocket and stably accommodate the BS/U2 duplex. This work explains the structural basis for the splicing modulation activity of PB and related compounds, and reveals key interactions between SF3B and a common pharmacophore, providing a framework for future structure-based drug design.
- Research Group Macromolecular Crystallography, Max Planck Institute for Biophysical Chemistry, Am Fassberg 11, 37077 Göttingen, Germany.
Organizational Affiliation: