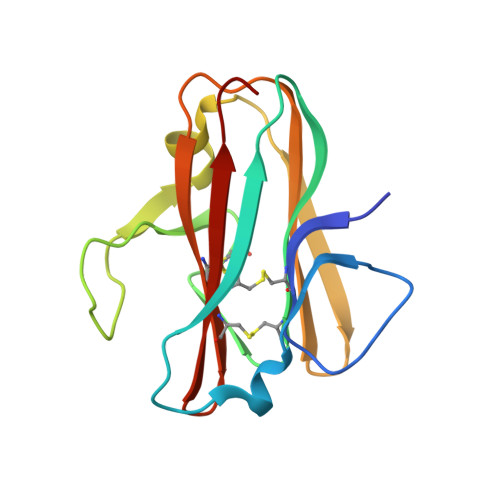
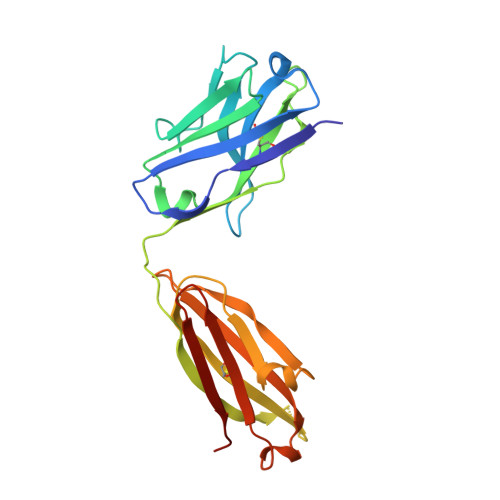
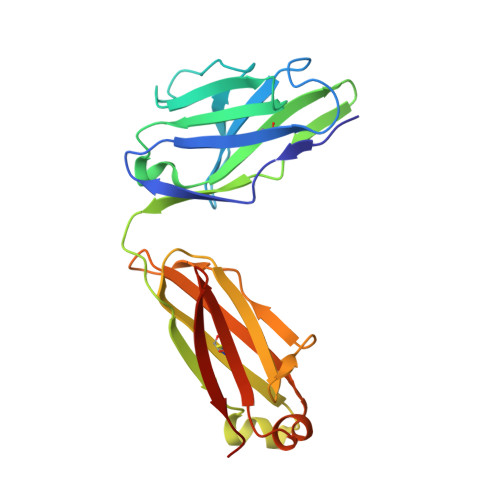
Structural delineation of potent transmission-blocking epitope I on malaria antigen Pfs48/45.
Kundu, P., Semesi, A., Jore, M.M., Morin, M.J., Price, V.L., Liang, A., Li, J., Miura, K., Sauerwein, R.W., King, C.R., Julien, J.P.(2018) Nat Commun 9: 4458-4458
- PubMed: 30367064
- DOI: https://doi.org/10.1038/s41467-018-06742-9
- Primary Citation of Related Structures:
6E62, 6E63, 6E64, 6E65 - PubMed Abstract:
Interventions that can block the transmission of malaria-causing Plasmodium falciparum (Pf) between the human host and Anopheles vector have the potential to reduce the incidence of malaria. Pfs48/45 is a gametocyte surface protein critical for parasite development and transmission, and its targeting by monoclonal antibody (mAb) 85RF45.1 leads to the potent reduction of parasite transmission. Here, we reveal how the Pfs48/45 6C domain adopts a (SAG1)-related-sequence (SRS) fold. We structurally delineate potent epitope I and show how mAb 85RF45.1 recognizes an electronegative surface with nanomolar affinity. Analysis of Pfs48/45 sequences reveals that polymorphisms are rare for residues involved at the binding interface. Humanization of rat-derived mAb 85RF45.1 conserved the mode of recognition and activity of the parental antibody, while also improving its thermostability. Our work has implications for the development of transmission-blocking interventions, both through improving vaccine designs and the testing of passive delivery of mAbs in humans.
- Program in Molecular Medicine, The Hospital for Sick Children Research Institute, Toronto, M5G 0A4, ON, Canada.
Organizational Affiliation: