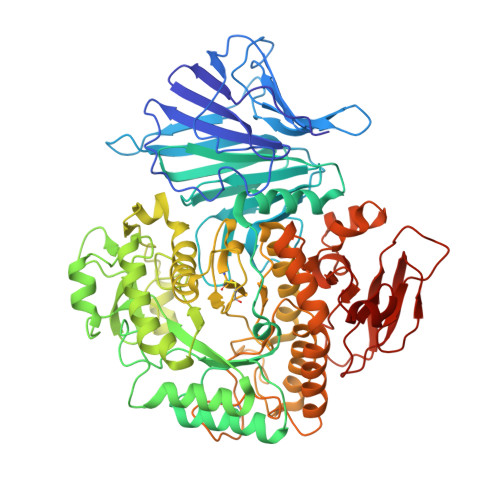
Crystal Structure of alpha-Xylosidase fromAspergillus nigerin Complex with a Hydrolyzed Xyloglucan Product and New Insights in Accurately Predicting Substrate Specificities of GH31 Family Glycosidases.
Cao, H., Walton, J.D., Brumm, P., Phillips Jr., G.N.(2020) ACS Sustain Chem Eng 8: 2540-2547
- PubMed: 32161692
- DOI: https://doi.org/10.1021/acssuschemeng.9b07073
- Primary Citation of Related Structures:
6DRU - PubMed Abstract:
Glycoside hydrolase family 31 (GH31) enzymes show both highly conserved folds and catalytic residues. Yet different members of GH31 show very different substrate specificities, and it is not obvious how these specificities arise from the protein sequences. The fungal α-xylosidase, AxlA, was originally isolated from a commercial enzyme mixture secreted by Aspergillus niger and was reported to have potential as a catalytic component in biomass deconstruction in the biofuel industry. We report here the crystal structure of AxlA in complex with its catalytic product, a hydrolyzed xyloglucan oligosaccharide. On the basis of our new structure, we provide the structural basis for AxlA's role in xyloglucan utilization and, more importantly, a new procedure to predict and differentiate C5 vs C6 sugar specific activities based on protein sequences of the functionally diverse GH31 family enzymes.
- BioSciences at Rice and Department of Chemistry, Rice University, Houston, Texas 77251, United States.
Organizational Affiliation: