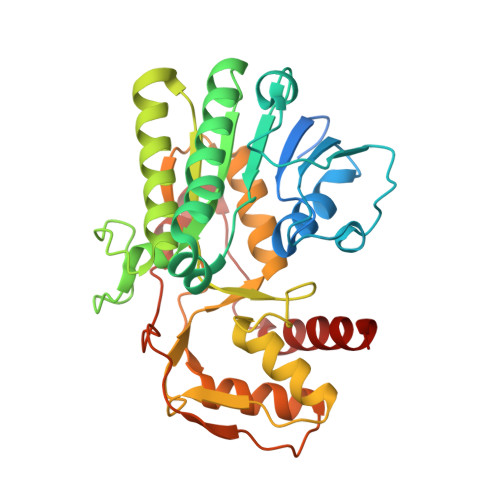
Structural determination of archaeal UDP-N-acetylglucosamine 4-epimerase from Methanobrevibacter ruminantium M1 in complex with the bacterial cell wall intermediate UDP-N-acetylmuramic acid.
Carbone, V., Schofield, L.R., Sang, C., Sutherland-Smith, A.J., Ronimus, R.S.(2018) Proteins 86: 1306-1312
- PubMed: 30242905
- DOI: https://doi.org/10.1002/prot.25606
- Primary Citation of Related Structures:
6DNT - PubMed Abstract:
The crystal structure of UDP-N-acetylglucosamine 4-epimerase (UDP-GlcNAc 4-epimerase; WbpP; EC 5.1.3.7), from the archaeal methanogen Methanobrevibacter ruminantium strain M1, was determined to a resolution of 1.65 Å. The structure, with a single monomer in the crystallographic asymmetric unit, contained a conserved N-terminal Rossmann-fold for nucleotide binding and an active site positioned in the C-terminus. UDP-GlcNAc 4-epimerase is a member of the short-chain dehydrogenases/reductases superfamily, sharing sequence motifs and structural elements characteristic of this family of oxidoreductases and bacterial 4-epimerases. The protein was co-crystallized with coenzyme NADH and UDP-N-acetylmuramic acid, the latter an unintended inclusion and well known product of the bacterial enzyme MurB and a critical intermediate for bacterial cell wall synthesis. This is a non-native UDP sugar amongst archaea and was most likely incorporated from the E. coli expression host during purification of the recombinant enzyme.
- AgResearch Limited, Grasslands Research Centre, Palmerston North, New Zealand.
Organizational Affiliation: